[English] 日本語
 Yorodumi
Yorodumi- EMDB-13161: Cryo EM structure of bison NHA2 in detergent and N-terminal exten... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13161 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo EM structure of bison NHA2 in detergent and N-terminal extension helix | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane protein Sodium proton transporter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationlithium:proton antiporter activity / lithium ion transport / positive regulation of osteoclast development / sperm principal piece / sodium ion homeostasis / sodium:proton antiporter activity / regulation of insulin secretion involved in cellular response to glucose stimulus / clathrin-dependent endocytosis / flagellated sperm motility / sodium ion transport ...lithium:proton antiporter activity / lithium ion transport / positive regulation of osteoclast development / sperm principal piece / sodium ion homeostasis / sodium:proton antiporter activity / regulation of insulin secretion involved in cellular response to glucose stimulus / clathrin-dependent endocytosis / flagellated sperm motility / sodium ion transport / recycling endosome / mitochondrial membrane / recycling endosome membrane / synaptic vesicle membrane / basolateral plasma membrane / endosome membrane / apical plasma membrane / lysosomal membrane / metal ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Bison bison (American bison) Bison bison (American bison) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.15 Å | |||||||||
 Authors Authors | Matsuoka R / Fudim R | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structure, mechanism and lipid-mediated remodeling of the mammalian Na/H exchanger NHA2. Authors: Rei Matsuoka / Roman Fudim / Sukkyeong Jung / Chenou Zhang / Andre Bazzone / Yurie Chatzikyriakidou / Carol V Robinson / Norimichi Nomura / So Iwata / Michael Landreh / Laura Orellana / ...Authors: Rei Matsuoka / Roman Fudim / Sukkyeong Jung / Chenou Zhang / Andre Bazzone / Yurie Chatzikyriakidou / Carol V Robinson / Norimichi Nomura / So Iwata / Michael Landreh / Laura Orellana / Oliver Beckstein / David Drew /      Abstract: The Na/H exchanger SLC9B2, also known as NHA2, correlates with the long-sought-after Na/Li exchanger linked to the pathogenesis of diabetes mellitus and essential hypertension in humans. Despite the ...The Na/H exchanger SLC9B2, also known as NHA2, correlates with the long-sought-after Na/Li exchanger linked to the pathogenesis of diabetes mellitus and essential hypertension in humans. Despite the functional importance of NHA2, structural information and the molecular basis for its ion-exchange mechanism have been lacking. Here we report the cryo-EM structures of bison NHA2 in detergent and in nanodiscs, at 3.0 and 3.5 Å resolution, respectively. The bison NHA2 structure, together with solid-state membrane-based electrophysiology, establishes the molecular basis for electroneutral ion exchange. NHA2 consists of 14 transmembrane (TM) segments, rather than the 13 TMs previously observed in mammalian Na/H exchangers (NHEs) and related bacterial antiporters. The additional N-terminal helix in NHA2 forms a unique homodimer interface with a large intracellular gap between the protomers, which closes in the presence of phosphoinositol lipids. We propose that the additional N-terminal helix has evolved as a lipid-mediated remodeling switch for the regulation of NHA2 activity. #1:  Journal: Nat.Struct.Mol.Biol. / Year: 2022 Journal: Nat.Struct.Mol.Biol. / Year: 2022Title: Structure, mechanism and lipid-mediated remodeling of the mammalian Na+/H+ exchanger NHA2 Authors: Matsuoka R / Fudim F / Jung S / Drew F | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13161.map.gz emd_13161.map.gz | 26.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13161-v30.xml emd-13161-v30.xml emd-13161.xml emd-13161.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13161.png emd_13161.png | 79 KB | ||
| Masks |  emd_13161_msk_1.map emd_13161_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13161.cif.gz emd-13161.cif.gz | 6.8 KB | ||
| Others |  emd_13161_half_map_1.map.gz emd_13161_half_map_1.map.gz emd_13161_half_map_2.map.gz emd_13161_half_map_2.map.gz | 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13161 http://ftp.pdbj.org/pub/emdb/structures/EMD-13161 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13161 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13161 | HTTPS FTP |
-Validation report
| Summary document |  emd_13161_validation.pdf.gz emd_13161_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13161_full_validation.pdf.gz emd_13161_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_13161_validation.xml.gz emd_13161_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_13161_validation.cif.gz emd_13161_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13161 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13161 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13161 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13161 | HTTPS FTP |
-Related structure data
| Related structure data |  7p1iMC  7p1jC  7p1kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13161.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13161.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0375 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
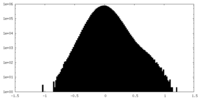
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13161_msk_1.map emd_13161_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
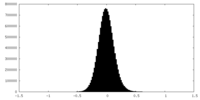
| Density Histograms |
-Half map: #2
| File | emd_13161_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
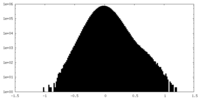
| Density Histograms |
-Half map: #1
| File | emd_13161_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
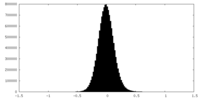
| Density Histograms |
- Sample components
Sample components
-Entire : detergent structure
| Entire | Name: detergent structure |
|---|---|
| Components |
|
-Supramolecule #1: detergent structure
| Supramolecule | Name: detergent structure / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Bison bison (American bison) Bison bison (American bison) |
| Molecular weight | Theoretical: 57373 kDa/nm |
-Macromolecule #1: mitochondrial sodium/hydrogen exchanger 9B2
| Macromolecule | Name: mitochondrial sodium/hydrogen exchanger 9B2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bison bison (American bison) Bison bison (American bison) |
| Molecular weight | Theoretical: 57.419203 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRNQDKRAAH KDSEPSTEVN HTASSYQGRQ QETGMNLRGI DGNEPTEGSN LLNNNEKMQG TPAEPNHLQR RRQIHACPPR GLLARVITN VTMVILLWAV VWSVTGSECL PGGNLFGIIM LFYCAIIGGK LFGLIKLPTL PPLPPLLGML LAGFLIRNVP V ISDNIQIK ...String: MRNQDKRAAH KDSEPSTEVN HTASSYQGRQ QETGMNLRGI DGNEPTEGSN LLNNNEKMQG TPAEPNHLQR RRQIHACPPR GLLARVITN VTMVILLWAV VWSVTGSECL PGGNLFGIIM LFYCAIIGGK LFGLIKLPTL PPLPPLLGML LAGFLIRNVP V ISDNIQIK HKWSSALRSI ALSVILVRAG LGLDSNALKK LKGVCVRLSL GPCLIEACTS AVLAYFLMGL PWQWGFMLGF VL GAVSPAV VVPSMLLLQE GGYGVEKGIP TLLMAAGSFD DILAITGFNT CLGMAFSTGS TVFNVLKGVL EVIIGVVTGL VLG FFIQYF PSSDQDNLVW KRAFLVLGLS VLAVFSSTYF GFPGSGGLCT LVTAFLAGRG WASTKTDVEK VIAVAWDIFQ PLLF GLIGA EVLITALRPE TIGLCVATLG IAVLIRILVT YLMVCFAGFN IKEKIFISFA WLPKATVQAA IGSVALDTAR SHGEK QLEG YGMDVLTVAF LSIIITAPVG SLLIGLLGPR LLQKAEQNKD EEDQGETSIQ V UniProtKB: Sodium/hydrogen exchanger 9B2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)