[English] 日本語
 Yorodumi
Yorodumi- EMDB-13157: Cryo-EM density of caspase-3 cleaved rXKR9 in complex with a sybo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13157 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM density of caspase-3 cleaved rXKR9 in complex with a sybody at 4.3A | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | small membrane protein / in complex with sybody / apoptotic lipid scrambling / MEMBRANE PROTEIN | |||||||||
| Function / homology | XK-related protein / : / XK-related protein / membrane / plasma membrane / XK-related protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
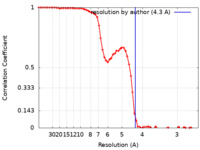
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Straub MS / Sawicka M | |||||||||
| Funding support |  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Cryo-EM structures of the caspase-activated protein XKR9 involved in apoptotic lipid scrambling. Authors: Monique S Straub / Carolina Alvadia / Marta Sawicka / Raimund Dutzler /  Abstract: The exposure of the negatively charged lipid phosphatidylserine on the cell surface, catalyzed by lipid scramblases, is an important signal for the clearance of apoptotic cells by macrophages. The ...The exposure of the negatively charged lipid phosphatidylserine on the cell surface, catalyzed by lipid scramblases, is an important signal for the clearance of apoptotic cells by macrophages. The protein XKR9 is a member of a conserved family that has been associated with apoptotic lipid scrambling. Here, we describe structures of full-length and caspase-treated XKR9 from in complex with a synthetic nanobody determined by cryo-electron microscopy. The 43 kDa monomeric membrane protein can be divided into two structurally related repeats, each containing four membrane-spanning segments and a helix that is partly inserted into the lipid bilayer. In the full-length protein, the C-terminus interacts with a hydrophobic pocket located at the intracellular side acting as an inhibitor of protein function. Cleavage by caspase-3 at a specific site releases 16 residues of the C-terminus, thus making the pocket accessible to the cytoplasm. Collectively, the work has revealed the unknown architecture of the XKR family and has provided initial insight into its activation by caspases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
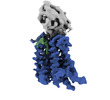
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13157.map.gz emd_13157.map.gz | 20.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13157-v30.xml emd-13157-v30.xml emd-13157.xml emd-13157.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13157_fsc.xml emd_13157_fsc.xml | 8.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_13157.png emd_13157.png | 348.4 KB | ||
| Masks |  emd_13157_msk_1.map emd_13157_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13157.cif.gz emd-13157.cif.gz | 6.1 KB | ||
| Others |  emd_13157_half_map_1.map.gz emd_13157_half_map_1.map.gz emd_13157_half_map_2.map.gz emd_13157_half_map_2.map.gz | 20.5 MB 20.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13157 http://ftp.pdbj.org/pub/emdb/structures/EMD-13157 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13157 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13157 | HTTPS FTP |
-Related structure data
| Related structure data |  7p16MC  7p14C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13157.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13157.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
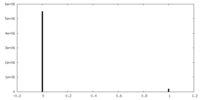
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.302 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
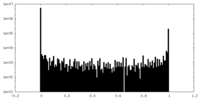
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13157_msk_1.map emd_13157_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
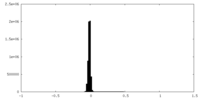
| Density Histograms |
-Half map: #2
| File | emd_13157_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
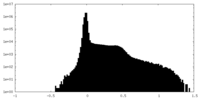
| Density Histograms |
-Half map: #1
| File | emd_13157_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : caspase-3 cleaved rXKR9 with sybody
| Entire | Name: caspase-3 cleaved rXKR9 with sybody |
|---|---|
| Components |
|
-Supramolecule #1: caspase-3 cleaved rXKR9 with sybody
| Supramolecule | Name: caspase-3 cleaved rXKR9 with sybody / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17 KDa |
-Supramolecule #2: rXKR9
| Supramolecule | Name: rXKR9 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Sybody
| Supramolecule | Name: Sybody / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: XK-related protein
| Macromolecule | Name: XK-related protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.563059 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKYTICNFMM SVLGIIIYVT DLVADIVLTV RYFYDGQYVF GVLTLSFVLC GTLIVHCFSY SWLKDDLKKA GGENEHYFLL LHCLQGGVF TRYWFVLRTG YHVVFKHSHR TSNFMEEQTD PHKEAIDMAT DLSMLRLFET YLEGCPQLIL QLYAFLERGQ A NFSQYMVI ...String: MKYTICNFMM SVLGIIIYVT DLVADIVLTV RYFYDGQYVF GVLTLSFVLC GTLIVHCFSY SWLKDDLKKA GGENEHYFLL LHCLQGGVF TRYWFVLRTG YHVVFKHSHR TSNFMEEQTD PHKEAIDMAT DLSMLRLFET YLEGCPQLIL QLYAFLERGQ A NFSQYMVI MVSCCAISWS TVDYQIALRK SLPDKNLLRG FWPKLTYLFY KLFTLLSWML SVVLLLFVDV RTVLLLLLFL WT VGFIWAF INHTQFCNSL SMEFLYRLVV GFILVFTFFN IKGQNTKCPM SCYYTVRVLG TLGILTVFWI YPLSIFNSDY FIP ISATIV LSLLFGIIFL GVYYGTYHPN INAGTQHDEP DGKAPQRDCR IRYFLMDA UniProtKB: XK-related protein |
-Macromolecule #2: Sybody
| Macromolecule | Name: Sybody / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 13.675293 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SQVQLVESGG GSVQAGGSLR LSCAASGNIA DIYYLGWFRQ APGKEREGVA ALITYNGRTY YADSVKGRFT VSLDNAKNTV YLQMNSLKP EDTALYYCAA AYNGLIAAPL KVTRYWYWGQ GTQVTVS |
-Macromolecule #3: DIUNDECYL PHOSPHATIDYL CHOLINE
| Macromolecule | Name: DIUNDECYL PHOSPHATIDYL CHOLINE / type: ligand / ID: 3 / Number of copies: 1 / Formula: PLC |
|---|---|
| Molecular weight | Theoretical: 622.834 Da |
| Chemical component information |  ChemComp-PLC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 14929 / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7p16: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)