[English] 日本語
 Yorodumi
Yorodumi- EMDB-1304: Limulus polyphemus hemocyanin: 10 A cryo-EM structure, sequence a... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1304 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Limulus polyphemus hemocyanin: 10 A cryo-EM structure, sequence analysis, molecular modelling and rigid-body fitting reveal the interfaces between the eight hexamers. | |||||||||
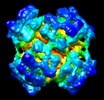
 Map data Map data | This is a map of the 8x6mer hemocyanin from the arthropod Limulus polyphemus. Mass correlated threshold: 0.001. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Limulus polyphemus (Atlantic horseshoe crab) Limulus polyphemus (Atlantic horseshoe crab) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 9.6 Å | |||||||||
 Authors Authors | Martin AG / Depoix F / Stohr M / Meissner U / Hagner-Holler S / Hammouti K / Burmester T / Heyd J / Wriggers W / Markl J | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2007 Journal: J Mol Biol / Year: 2007Title: Limulus polyphemus hemocyanin: 10 A cryo-EM structure, sequence analysis, molecular modelling and rigid-body fitting reveal the interfaces between the eight hexamers. Authors: Andreas G Martin / Frank Depoix / Michael Stohr / Ulrich Meissner / Silke Hagner-Holler / Kada Hammouti / Thorsten Burmester / Jochen Heyd / Willy Wriggers / Jürgen Markl /  Abstract: The blue copper protein hemocyanin from the horseshoe crab Limulus polyphemus is among the largest respiratory proteins found in nature (3.5 MDa) and exhibits a highly cooperative oxygen binding. Its ...The blue copper protein hemocyanin from the horseshoe crab Limulus polyphemus is among the largest respiratory proteins found in nature (3.5 MDa) and exhibits a highly cooperative oxygen binding. Its 48 subunits are arranged as eight hexamers (1x6mers) that form the native 8x6mer in a nested hierarchy of 2x6mers and 4x6mers. This quaternary structure is established by eight subunit types (termed I, IIA, II, IIIA, IIIB, IV, V, and VI), of which only type II has been sequenced. Crystal structures of the 1x6mer are available, but for the 8x6mer only a 40 A 3D reconstruction exists. Consequently, the structural parameters of the 8x6mer are not firmly established, and the molecular interfaces between the eight hexamers are still to be defined. This, however, is crucial for understanding how allosteric transitions are mediated between the different levels of hierarchy. Here, we show the 10 A structure (FSC(1/2-bit) criterion) of the oxygenated 8x6mer from cryo-electron microscopy (cryo-EM) and single-particle analysis. Moreover, we show its molecular model as obtained by DNA sequencing of subunits II, IIIA, IV and VI, and molecular modelling and rigid-body fitting of all subunit types. Remarkably, the latter enabled us to improve the resolution of the cryo-EM structure from 11 A to the final 10 A. The 10 A structure allows firm assessment of various structural parameters of the 8x6mer, the 4x6mer and the 2x6mer, and reveals a total of 46 inter-hexamer bridges. These group as 11 types of interface: four at the 2x6mer level (II-II, II-IV, V-VI, IV-VI), three form the 4x6mer (V-V, V-VI, VI-IIIB/IV/V), and four are required to assemble the 8x6mer (IIIA-IIIA, IIIA-IIIB, II-IV, IV-IV). The molecular model shows the amino acid residues involved, and reveals that several of the interfaces are intriguingly histidine-rich and likely to transfer allosteric signals between the different levels of the nested hierarchy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1304.map.gz emd_1304.map.gz | 20.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1304-v30.xml emd-1304-v30.xml emd-1304.xml emd-1304.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1304.png EMD-1304.png | 273 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1304 http://ftp.pdbj.org/pub/emdb/structures/EMD-1304 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1304 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1304 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1304.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1304.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of the 8x6mer hemocyanin from the arthropod Limulus polyphemus. Mass correlated threshold: 0.001. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 8x6mer arthropod hemocyanin
| Entire | Name: 8x6mer arthropod hemocyanin |
|---|---|
| Components |
|
-Supramolecule #1000: 8x6mer arthropod hemocyanin
| Supramolecule | Name: 8x6mer arthropod hemocyanin / type: sample / ID: 1000 / Oligomeric state: hetero-oligomeric 8x6mer / Number unique components: 8 |
|---|---|
| Molecular weight | Theoretical: 3.5 MDa / Method: sequence analysis |
-Macromolecule #1: subunit type I
| Macromolecule | Name: subunit type I / type: protein_or_peptide / ID: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Limulus polyphemus (Atlantic horseshoe crab) Limulus polyphemus (Atlantic horseshoe crab) |
-Macromolecule #2: subunit type IIA
| Macromolecule | Name: subunit type IIA / type: protein_or_peptide / ID: 2 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Limulus polyphemus (Atlantic horseshoe crab) Limulus polyphemus (Atlantic horseshoe crab) |
-Macromolecule #3: subunit type II
| Macromolecule | Name: subunit type II / type: protein_or_peptide / ID: 3 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Limulus polyphemus (Atlantic horseshoe crab) Limulus polyphemus (Atlantic horseshoe crab) |
-Macromolecule #4: subunit type IIIA
| Macromolecule | Name: subunit type IIIA / type: protein_or_peptide / ID: 4 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Limulus polyphemus (Atlantic horseshoe crab) Limulus polyphemus (Atlantic horseshoe crab) |
-Macromolecule #5: subunit type IIIB
| Macromolecule | Name: subunit type IIIB / type: protein_or_peptide / ID: 5 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Limulus polyphemus (Atlantic horseshoe crab) Limulus polyphemus (Atlantic horseshoe crab) |
-Macromolecule #6: subunit type IV
| Macromolecule | Name: subunit type IV / type: protein_or_peptide / ID: 6 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Limulus polyphemus (Atlantic horseshoe crab) Limulus polyphemus (Atlantic horseshoe crab) |
-Macromolecule #7: subunit type V
| Macromolecule | Name: subunit type V / type: protein_or_peptide / ID: 7 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Limulus polyphemus (Atlantic horseshoe crab) Limulus polyphemus (Atlantic horseshoe crab) |
-Macromolecule #8: subunit type VI
| Macromolecule | Name: subunit type VI / type: protein_or_peptide / ID: 8 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Limulus polyphemus (Atlantic horseshoe crab) Limulus polyphemus (Atlantic horseshoe crab) |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.8 / Details: 100mM Tris-HCL,10mM MgCl2, 10mM CaCl2 |
| Staining | Type: NEGATIVE / Details: cryo-EM, no stain |
| Grid | Details: 400 mesh copper |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 86 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: self constructed plunger. Vitrification carried out in 25% oxygen - 75% nitrogen atmosphere Method: Single side blotting and rapid plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Temperature | Average: 86 K |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: PRIMESCAN / Digitization - Sampling interval: 1.8 µm / Number real images: 29 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 59000 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 1.2 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder: Gatan single-tilt cryoholder / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were selected using the automatic selection program boxer. |
|---|---|
| CTF correction | Details: CTFFIND3 and TRANSFER, IMAGIC5 |
| Final reconstruction | Applied symmetry - Point group: D2 (2x2 fold dihedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.6 Å / Resolution method: OTHER / Software - Name: IMAGIC-5 / Number images used: 12069 |
| Final two d classification | Number classes: 1140 |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















