[English] 日本語
 Yorodumi
Yorodumi- EMDB-13020: CryoEM structure of DNA Polymerase alpha - primase bound to SARS ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13020 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
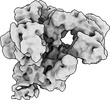
| Title | CryoEM structure of DNA Polymerase alpha - primase bound to SARS CoV nsp1 | |||||||||
 Map data Map data | 3D refinement map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA polymerase / Primase / viral protein / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA primase AEP / ribonucleotide binding / DNA replication initiation / DNA/RNA hybrid binding / Inhibition of replication initiation of damaged DNA by RB1/E2F1 / regulation of type I interferon production / Telomere C-strand synthesis initiation / alpha DNA polymerase:primase complex / Assembly of the SARS-CoV-1 Replication-Transcription Complex (RTC) / Maturation of replicase proteins ...DNA primase AEP / ribonucleotide binding / DNA replication initiation / DNA/RNA hybrid binding / Inhibition of replication initiation of damaged DNA by RB1/E2F1 / regulation of type I interferon production / Telomere C-strand synthesis initiation / alpha DNA polymerase:primase complex / Assembly of the SARS-CoV-1 Replication-Transcription Complex (RTC) / Maturation of replicase proteins / Transcription of SARS-CoV-1 sgRNAs / Polymerase switching / Processive synthesis on the lagging strand / Removal of the Flap Intermediate / lagging strand elongation / mitotic DNA replication initiation / Translation of Replicase and Assembly of the Replication Transcription Complex / DNA replication, synthesis of primer / Polymerase switching on the C-strand of the telomere / K48-linked deubiquitinase activity / Replication of the SARS-CoV-1 genome / K63-linked deubiquitinase activity / host cell endoplasmic reticulum / DNA strand elongation involved in DNA replication / DNA synthesis involved in DNA repair / G1/S-Specific Transcription / leading strand elongation / SARS-CoV-1 modulates host translation machinery / DNA replication origin binding / DNA replication initiation / Activation of the pre-replicative complex / viral genome replication / methyltransferase activity / Defective pyroptosis / double-strand break repair via nonhomologous end joining / nuclear matrix / protein import into nucleus / SARS-CoV-1 activates/modulates innate immune responses / DNA-directed RNA polymerase activity / nuclear envelope / single-stranded DNA binding / 4 iron, 4 sulfur cluster binding / endonuclease activity / double membrane vesicle viral factory outer membrane / SARS coronavirus main proteinase / host cell endosome / symbiont-mediated degradation of host mRNA / methylation / mRNA guanylyltransferase / symbiont-mediated suppression of host ISG15-protein conjugation / G-quadruplex RNA binding / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / omega peptidase activity / DNA-directed DNA polymerase / host cell Golgi apparatus / symbiont-mediated perturbation of host ubiquitin-like protein modification / DNA-directed DNA polymerase activity / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / DNA replication / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / single-stranded RNA binding / regulation of autophagy / viral protein processing / lyase activity / ciliary basal body / host cell perinuclear region of cytoplasm / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont-mediated activation of host autophagy / cysteine-type endopeptidase activity / DNA repair / nucleotide binding / intracellular membrane-bounded organelle / RNA-directed RNA polymerase activity / chromatin binding / protein kinase binding / chromatin / nucleolus / magnesium ion binding / proteolysis / DNA binding / zinc ion binding / nucleoplasm / metal ion binding / identical protein binding / nucleus / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Severe acute respiratory syndrome coronavirus Severe acute respiratory syndrome coronavirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.12 Å | |||||||||
 Authors Authors | Kilkenny TJ / Pellegrini L | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Protein Sci / Year: 2022 Journal: Protein Sci / Year: 2022Title: Structural basis for the interaction of SARS-CoV-2 virulence factor nsp1 with DNA polymerase α-primase. Authors: Mairi L Kilkenny / Charlotte E Veale / Amir Guppy / Steven W Hardwick / Dimitri Y Chirgadze / Neil J Rzechorzek / Joseph D Maman / Luca Pellegrini /  Abstract: The molecular mechanisms that drive the infection by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-the causative agent of coronavirus disease 2019 (COVID-19)-are under intense ...The molecular mechanisms that drive the infection by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-the causative agent of coronavirus disease 2019 (COVID-19)-are under intense current scrutiny to understand how the virus operates and to uncover ways in which the disease can be prevented or alleviated. Recent proteomic screens of the interactions between viral and host proteins have identified the human proteins targeted by SARS-CoV-2. The DNA polymerase α (Pol α)-primase complex or primosome-responsible for initiating DNA synthesis during genomic duplication-was identified as a target of nonstructural protein 1 (nsp1), a major virulence factor in the SARS-CoV-2 infection. Here, we validate the published reports of the interaction of nsp1 with the primosome by demonstrating direct binding with purified recombinant components and providing a biochemical characterization of their interaction. Furthermore, we provide a structural basis for the interaction by elucidating the cryo-electron microscopy structure of nsp1 bound to the primosome. Our findings provide biochemical evidence for the reported targeting of Pol α by the virulence factor nsp1 and suggest that SARS-CoV-2 interferes with Pol α's putative role in the immune response during the viral infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13020.map.gz emd_13020.map.gz | 11.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13020-v30.xml emd-13020-v30.xml emd-13020.xml emd-13020.xml | 29 KB 29 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13020.png emd_13020.png | 114.3 KB | ||
| Filedesc metadata |  emd-13020.cif.gz emd-13020.cif.gz | 8.2 KB | ||
| Others |  emd_13020_additional_1.map.gz emd_13020_additional_1.map.gz emd_13020_half_map_1.map.gz emd_13020_half_map_1.map.gz emd_13020_half_map_2.map.gz emd_13020_half_map_2.map.gz | 14.6 MB 11.9 MB 11.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13020 http://ftp.pdbj.org/pub/emdb/structures/EMD-13020 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13020 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13020 | HTTPS FTP |
-Validation report
| Summary document |  emd_13020_validation.pdf.gz emd_13020_validation.pdf.gz | 740.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13020_full_validation.pdf.gz emd_13020_full_validation.pdf.gz | 739.7 KB | Display | |
| Data in XML |  emd_13020_validation.xml.gz emd_13020_validation.xml.gz | 9.3 KB | Display | |
| Data in CIF |  emd_13020_validation.cif.gz emd_13020_validation.cif.gz | 10.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13020 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13020 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13020 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13020 | HTTPS FTP |
-Related structure data
| Related structure data |  7oplMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13020.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13020.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D refinement map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.314 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sharpened map, calculated from the two half maps...
| File | emd_13020_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map, calculated from the two half maps using local, anisotropic sharpening in Phenix. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map #1
| File | emd_13020_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map #2
| File | emd_13020_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of DNA polymerase alpha - primase bound to SARS COV-2 nsp1
| Entire | Name: Complex of DNA polymerase alpha - primase bound to SARS COV-2 nsp1 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of DNA polymerase alpha - primase bound to SARS COV-2 nsp1
| Supramolecule | Name: Complex of DNA polymerase alpha - primase bound to SARS COV-2 nsp1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 310 KDa |
-Macromolecule #1: DNA polymerase alpha catalytic subunit
| Macromolecule | Name: DNA polymerase alpha catalytic subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 133.702562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAWSHPQFE KGGGSGGGSG GGSWSHPQFE KLEVLFQGPE FGADEEQVFH FYWLDAYEDQ YNQPGVVFLF GKVWIESAET HVSCCVMVK NIERTLYFLP REMKIDLNTG KETGTPISMK DVYEEFDEKI ATKYKIMKFK SKPVEKNYAF EIPDVPEKSE Y LEVKYSAE ...String: MSAWSHPQFE KGGGSGGGSG GGSWSHPQFE KLEVLFQGPE FGADEEQVFH FYWLDAYEDQ YNQPGVVFLF GKVWIESAET HVSCCVMVK NIERTLYFLP REMKIDLNTG KETGTPISMK DVYEEFDEKI ATKYKIMKFK SKPVEKNYAF EIPDVPEKSE Y LEVKYSAE MPQLPQDLKG ETFSHVFGTN TSSLELFLMN RKIKGPCWLE VKSPQLLNQP VSWCKVEAMA LKPDLVNVIK DV SPPPLVV MAFSMKTMQN AKNHQNEIIA MAALVHHSFA LDKAAPKPPF QSHFCVVSKP KDCIFPYAFK EVIEKKNVKV EVA ATERTL LGFFLAKVHK IDPDIIVGHN IYGFELEVLL QRINVCKAPH WSKIGRLKRS NMPKLGGRSG FGERNATCGR MICD VEISA KELIRCKSYH LSELVQQILK TERVVIPMEN IQNMYSESSQ LLYLLEHTWK DAKFILQIMC ELNVLPLALQ ITNIA GNIM SRTLMGGRSE RNEFLLLHAF YENNYIVPDK QIFRKPQQKL GDEDEEIDGD TNKYKKGRKK AAYAGGLVLD PKVGFY DKF ILLLDFNSLY PSIIQEFNIC FTTVQRVASE AQKVTEDGEQ EQIPELPDPS LEMGILPREI RKLVERRKQV KQLMKQQ DL NPDLILQYDI RQKALKLTAN SMYGCLGFSY SRFYAKPLAA LVTYKGREIL MHTKEMVQKM NLEVIYGDTD SIMINTNS T NLEEVFKLGN KVKSEVNKLY KLLEIDIDGV FKSLLLLKKK KYAALVVEPT SDGNYVTKQE LKGLDIVRRD WCDLAKDTG NFVIGQILSD QSRDTIVENI QKRLIEIGEN VLNGSVPVSQ FEINKALTKD PQDYPDKKSL PHVHVALWIN SQGGRKVKAG DTVSYVICQ DGSNLTASQR AYAPEQLQKQ DNLTIDTQYY LAQQIHPVVA RICEPIDGID AVLIATWLGL DPTQFRVHHY H KDEENDAL LGGPAQLTDE EKYRDCERFK CPCPTCGTEN IYDNVFDGSG TDMEPSLYRC SNIDCKASPL TFTVQLSNKL IM DIRRFIK KYYDGWLICE EPTCRNRTRH LPLQFSRTGP LCPACMKATL QPEYSDKSLY TQLCFYRYIF DAECALEKLT TDH EKDKLK KQFFTPKVLQ DYRKLKNTAE QFLSRSGYSE VNLSKLFAGC AVKS UniProtKB: DNA polymerase alpha catalytic subunit |
-Macromolecule #2: DNA polymerase alpha subunit B
| Macromolecule | Name: DNA polymerase alpha subunit B / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.855434 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSFSPSAT PSQKYNSRSN RGEVVTSFGL AQGVSWSGRG GAGNISLKVL GCPEALTGSY KSMFQKLPDI REVLTCKIEE LGSELKEHY KIEAFTPLLA PAQEPVTLLG QIGCDSNGKL NNKSVILEGD REHSSGAQIP VDLSELKEYS LFPGQVVIME G INTTGRKL ...String: MGSSFSPSAT PSQKYNSRSN RGEVVTSFGL AQGVSWSGRG GAGNISLKVL GCPEALTGSY KSMFQKLPDI REVLTCKIEE LGSELKEHY KIEAFTPLLA PAQEPVTLLG QIGCDSNGKL NNKSVILEGD REHSSGAQIP VDLSELKEYS LFPGQVVIME G INTTGRKL VATKLYEGVP LPFYQPTEED ADFEQSMVLV ACGPYTTSDS ITYDPLLDLI AVINHDRPDV CILFGPFLDA KH EQVENCL LTSPFEDIFK QCLRTIIEGT RSSGSHLVFV PSLRDVHHEP VYPQPPFSYS DLSREDKKQV QFVSEPCSLS ING VIFGLT STDLLFHLGA EEISSSSGTS DRFSRILKHI LTQRSYYPLY PPQEDMAIDY ESFYVYAQLP VTPDVLIIPS ELRY FVKDV LGCVCVNPGR LTKGQVGGTF ARLYLRRPAA DGAERQSPCI AVQVVRI UniProtKB: DNA polymerase alpha subunit B |
-Macromolecule #3: DNA primase small subunit
| Macromolecule | Name: DNA primase small subunit / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.590801 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HGENLYFQGT SMETFDPTEL PELLKLYYRR LFPYSQYYRW LNYGGVIKNY FQHREFSFTL KDDIYIRYQS FNNQSDLEK EMQKMNPYKI DIGAVYSHRP NQHNTVKLGA FQAQEKELVF DIDMTDYDDV RRCCSSADIC PKCWTLMTMA I RIIDRALK ...String: MHHHHHHHHH HGENLYFQGT SMETFDPTEL PELLKLYYRR LFPYSQYYRW LNYGGVIKNY FQHREFSFTL KDDIYIRYQS FNNQSDLEK EMQKMNPYKI DIGAVYSHRP NQHNTVKLGA FQAQEKELVF DIDMTDYDDV RRCCSSADIC PKCWTLMTMA I RIIDRALK EDFGFKHRLW VYSGRRGVHC WVCDESVRKL SSAVRSGIVE YLSLVKGGQD VKKKVHLSEK IHPFIRKSIN II KKYFEEY ALVNQDILEN KESWDKILAL VPETIHDELQ QSFQKSHNSL QRWEHLKKVA SRYQNNIKND KYGPWLEWEI MLQ YCFPRL DINVSKGINH LLKSPFSVHP KTGRISVPID LQKVDQFDPF TVPTISFICR ELDAISTNEE EKEENEAESD VKHR TRDYK KTSLAPYVKV FEHFLENLDK SRKGELLKKS DLQKDF UniProtKB: DNA primase small subunit |
-Macromolecule #4: DNA primase large subunit
| Macromolecule | Name: DNA primase large subunit / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 58.890918 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEFSGRKWRK LRLAGDQRNA SYPHCLQFYL QPPSENISLI EFENLAIDRV KLLKSVENLG VSYVKGTEQY QSKLESELRK LKFSYRENL EDEYEPRRRD HISHFILRLA YCQSEELRRW FIQQEMDLLR FRFSILPKDK IQDFLKDSQL QFEAISDEEK T LREQEIVA ...String: MEFSGRKWRK LRLAGDQRNA SYPHCLQFYL QPPSENISLI EFENLAIDRV KLLKSVENLG VSYVKGTEQY QSKLESELRK LKFSYRENL EDEYEPRRRD HISHFILRLA YCQSEELRRW FIQQEMDLLR FRFSILPKDK IQDFLKDSQL QFEAISDEEK T LREQEIVA SSPSLSGLKL GFESIYKIPF ADALDLFRGR KVYLEDGFAY VPLKDIVAII LNEFRAKLSK ALALTARSLP AV QSDERLQ PLLNHLSHSY TGQDYSTQGN VGKISLDQID LLSTKSFPPC MRQLHKALRE NHHLRHGGRM QYGLFLKGIG LTL EQALQF WKQEFIKGKM DPDKFDKGYS YNIRHSFGKE GKRTDYTPFS CLKIILSNPP SQGDYHGCPF RHSDPELLKQ KLQS YKISP GGISQILDLV KGTHYQVACQ KYFEMIHNVD DCGFSLNHPN QFFCESQRIL NGGKDIKKEP IQPETPQPKP SVQKT KDAS SALASLNSSL EMDMEGLEDY FSEDS UniProtKB: DNA primase large subunit |
-Macromolecule #5: Non-structural protein 1
| Macromolecule | Name: Non-structural protein 1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Severe acute respiratory syndrome coronavirus Severe acute respiratory syndrome coronavirus |
| Molecular weight | Theoretical: 12.955852 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSMGHVQLSL PVLQVRDVLV RGFGDSVEEA LSEAREHLKN GTCGLVELEK GVLPQLEQPY VFIKRSDALS TNHGHKVVEL VAEMDGIQY GRSGITLGVL VPHVGETPIA YRNVLLRKNG UniProtKB: Replicase polyprotein 1a |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #7: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 7 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| ||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV | ||||||||
| Details | The nsp1 protein was added in 10-fold stoichiometric excess |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 2919 / Average exposure time: 1.31 sec. / Average electron dose: 46.91 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -0.0007 µm / Nominal defocus min: -0.0025 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||||||||||
| Output model |  PDB-7opl: |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)