Yorodumi
Yorodumi+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of undecameric SlyB from Escherichia coli K12 | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | OUTER MEMBRANE CHAPERONE / 2TM GLYCINE ZIPPER / OUTER MEMBRANE LIPOPROTEIN SLYB / LPS-LP BINDING PROTEIN / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology | : / Glycine zipper 2TM domain / Glycine zipper 2TM domain / outer membrane / Prokaryotic membrane lipoprotein lipid attachment site profile. / Outer membrane lipoprotein slyB Function and homology information Function and homology information | |||||||||||||||
| Biological species |  | |||||||||||||||
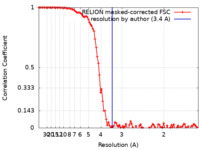
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||||||||
 Authors Authors | Nguyen VS / Remaut H | |||||||||||||||
| Funding support |  Belgium, 4 items Belgium, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: SlyB encapsulates outer membrane proteins in stress-induced lipid nanodomains Authors: Janssens A / Nguyen VS / Cecil AJ / Van der Verren SE / Timmerman E / Deghelt M / Pak AJ / Collet JF / Impens F / Remaut H | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12950.map.gz emd_12950.map.gz | 85.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12950-v30.xml emd-12950-v30.xml emd-12950.xml emd-12950.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12950_fsc.xml emd_12950_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_12950.png emd_12950.png | 174.4 KB | ||
| Filedesc metadata |  emd-12950.cif.gz emd-12950.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12950 http://ftp.pdbj.org/pub/emdb/structures/EMD-12950 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12950 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12950 | HTTPS FTP |
-Validation report
| Summary document |  emd_12950_validation.pdf.gz emd_12950_validation.pdf.gz | 584.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12950_full_validation.pdf.gz emd_12950_full_validation.pdf.gz | 583.6 KB | Display | |
| Data in XML |  emd_12950_validation.xml.gz emd_12950_validation.xml.gz | 11.6 KB | Display | |
| Data in CIF |  emd_12950_validation.cif.gz emd_12950_validation.cif.gz | 15.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12950 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12950 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12950 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12950 | HTTPS FTP |
-Related structure data
| Related structure data |  7ojgMC  7ojfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12950.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12950.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.784 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Outer membrane lipoprotein SlyB
| Entire | Name: Outer membrane lipoprotein SlyB |
|---|---|
| Components |
|
-Supramolecule #1: Outer membrane lipoprotein SlyB
| Supramolecule | Name: Outer membrane lipoprotein SlyB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Undecameric SlyB was purified from overexpression of SlyB-TEV-HIS in E. coli using using 2-step Ni-IMAC purification. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Outer membrane lipoprotein slyB
| Macromolecule | Name: Outer membrane lipoprotein slyB / type: protein_or_peptide / ID: 1 / Number of copies: 11 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.613606 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIKRVLVVSM VGLSLVGCVN NDTLSGDVYT ASEAKQVQNV SYGTIVNVRP VQIQGGDDSN VIGAIGGAVL GGFLGNTVGG GTGRSLATA AGAVAGGVAG QGVQSAMNKT QGVELEIRKD DGNTIMVVQK QGNTRFSPGQ RVVLASNGSQ VTVSPR UniProtKB: Outer membrane lipoprotein slyB |
-Macromolecule #2: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 2 / Number of copies: 33 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Macromolecule #3: GLYCEROL
| Macromolecule | Name: GLYCEROL / type: ligand / ID: 3 / Number of copies: 11 / Formula: GOL |
|---|---|
| Molecular weight | Theoretical: 92.094 Da |
| Chemical component information |  ChemComp-GOL: |
-Macromolecule #4: (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[[...
| Macromolecule | Name: (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(~{E},3~{R})-3-dodecanoyloxytetradec-5-enoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3- ...Name: (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(~{E},3~{R})-3-dodecanoyloxytetradec-5-enoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(~{E},3~{R})-3-oxidanyltetradec-11-enoyl]amino]-4-[(~{E},3~{R})-3-oxidanyltetradec-5-enoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(~{E},3~{R})-3-tetradecanoyloxytetradec-7-enoyl]oxy-oxan-2-yl]methoxy]-4,5-bis(oxidanyl)oxane-2-carboxylic acid type: ligand / ID: 4 / Number of copies: 11 / Formula: L8Z |
|---|---|
| Molecular weight | Theoretical: 2.010478 KDa |
| Chemical component information | 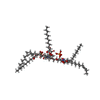 ChemComp-L8Z: |
-Macromolecule #5: 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE
| Macromolecule | Name: 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE type: ligand / ID: 5 / Number of copies: 11 / Formula: LPP |
|---|---|
| Molecular weight | Theoretical: 648.891 Da |
| Chemical component information |  ChemComp-LPP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.04 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: GRAPHENE OXIDE / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: GATAN CRYOPLUNGE 3 / Details: Back-blotting for 4 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 6352 / Average exposure time: 3.0 sec. / Average electron dose: 61.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.55 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 35 |
|---|---|
| Output model |  PDB-7ojg: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)