+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12893 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Drs2p-Cdc50p in the E1 state | ||||||||||||
 Map data Map data | Sharpened map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Lipid Flippase / P4 ATPase / trans-Golgi Network / Phosphatidylserine transport / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationCdc50p-Drs2p complex / actin cortical patch localization / aminophospholipid translocation / phosphatidylcholine flippase activity / Ion transport by P-type ATPases / post-Golgi vesicle-mediated transport / phosphatidylserine flippase activity / phospholipid-translocating ATPase complex / phosphatidylserine floppase activity / ATPase-coupled intramembrane lipid transporter activity ...Cdc50p-Drs2p complex / actin cortical patch localization / aminophospholipid translocation / phosphatidylcholine flippase activity / Ion transport by P-type ATPases / post-Golgi vesicle-mediated transport / phosphatidylserine flippase activity / phospholipid-translocating ATPase complex / phosphatidylserine floppase activity / ATPase-coupled intramembrane lipid transporter activity / phosphatidylethanolamine flippase activity / endocytic recycling / P-type phospholipid transporter / phosphatidylinositol-4-phosphate binding / retrograde transport, endosome to Golgi / phospholipid translocation / Neutrophil degranulation / intracellular protein transport / trans-Golgi network / endocytosis / late endosome membrane / endosome membrane / magnesium ion binding / endoplasmic reticulum / Golgi apparatus / ATP hydrolysis activity / ATP binding / plasma membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Timcenko M / Dieudonne T | ||||||||||||
| Funding support |  Denmark, Denmark,  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2021 Journal: J Mol Biol / Year: 2021Title: Structural Basis of Substrate-Independent Phosphorylation in a P4-ATPase Lipid Flippase. Authors: Milena Timcenko / Thibaud Dieudonné / Cédric Montigny / Thomas Boesen / Joseph A Lyons / Guillaume Lenoir / Poul Nissen /   Abstract: P4-ATPases define a eukaryotic subfamily of the P-type ATPases, and are responsible for the transverse flip of specific lipids from the extracellular or luminal leaflet to the cytosolic leaflet of ...P4-ATPases define a eukaryotic subfamily of the P-type ATPases, and are responsible for the transverse flip of specific lipids from the extracellular or luminal leaflet to the cytosolic leaflet of cell membranes. The enzymatic cycle of P-type ATPases is divided into autophosphorylation and dephosphorylation half-reactions. Unlike most other P-type ATPases, P4-ATPases transport their substrate during dephosphorylation only, i.e. the phosphorylation half-reaction is not associated with transport. To study the structural basis of the distinct mechanisms of P4-ATPases, we have determined cryo-EM structures of Drs2p-Cdc50p from Saccharomyces cerevisiae covering multiple intermediates of the cycle. We identify several structural motifs specific to Drs2p and P4-ATPases in general that decrease movements and flexibility of domains as compared to other P-type ATPases such as Na/K-ATPase or Ca-ATPase. These motifs include the linkers that connect the transmembrane region to the actuator (A) domain, which is responsible for dephosphorylation. Additionally, mutation of Tyr380, which interacts with conserved Asp340 of the distinct DGET dephosphorylation loop of P4-ATPases, highlights a functional role of these P4-ATPase specific motifs in the A-domain. Finally, the transmembrane (TM) domain, responsible for transport, also undergoes less extensive conformational changes, which is ensured both by a longer segment connecting TM helix 4 with the phosphorylation site, and possible stabilization by the auxiliary subunit Cdc50p. Collectively these adaptions in P4-ATPases are responsible for phosphorylation becoming transport-independent. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12893.map.gz emd_12893.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12893-v30.xml emd-12893-v30.xml emd-12893.xml emd-12893.xml | 25.6 KB 25.6 KB | Display Display |  EMDB header EMDB header |
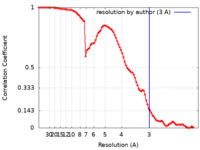
| FSC (resolution estimation) |  emd_12893_fsc.xml emd_12893_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_12893.png emd_12893.png | 101 KB | ||
| Filedesc metadata |  emd-12893.cif.gz emd-12893.cif.gz | 8.1 KB | ||
| Others |  emd_12893_half_map_1.map.gz emd_12893_half_map_1.map.gz emd_12893_half_map_2.map.gz emd_12893_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12893 http://ftp.pdbj.org/pub/emdb/structures/EMD-12893 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12893 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12893 | HTTPS FTP |
-Validation report
| Summary document |  emd_12893_validation.pdf.gz emd_12893_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12893_full_validation.pdf.gz emd_12893_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_12893_validation.xml.gz emd_12893_validation.xml.gz | 16.6 KB | Display | |
| Data in CIF |  emd_12893_validation.cif.gz emd_12893_validation.cif.gz | 21.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12893 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12893 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12893 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12893 | HTTPS FTP |
-Related structure data
| Related structure data |  7oh4MC  7oh5C  7oh6C  7oh7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12893.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12893.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0312 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half-map A
| File | emd_12893_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
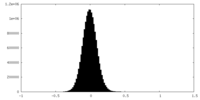
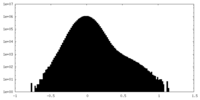
| Density Histograms |
-Half map: Half-map B
| File | emd_12893_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
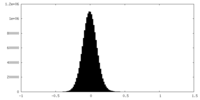
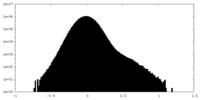
| Density Histograms |
- Sample components
Sample components
-Entire : Binary complex of Drs2p-Cdc50p with the regulatory lipid PI4P
| Entire | Name: Binary complex of Drs2p-Cdc50p with the regulatory lipid PI4P |
|---|---|
| Components |
|
-Supramolecule #1: Binary complex of Drs2p-Cdc50p with the regulatory lipid PI4P
| Supramolecule | Name: Binary complex of Drs2p-Cdc50p with the regulatory lipid PI4P type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: Drs2p
| Supramolecule | Name: Drs2p / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Cdc50p
| Supramolecule | Name: Cdc50p / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Probable phospholipid-transporting ATPase DRS2,Probable phospholi...
| Macromolecule | Name: Probable phospholipid-transporting ATPase DRS2,Probable phospholipid-transporting ATPase DRS2 type: protein_or_peptide / ID: 1 Details: Drs2p with a Thrombin cleavable C-terminal biotin acceptor domain (BAD) tag, and additional Thrombin cleavage site in the C-terminus.,Drs2p with a Thrombin cleavable C-terminal biotin ...Details: Drs2p with a Thrombin cleavable C-terminal biotin acceptor domain (BAD) tag, and additional Thrombin cleavage site in the C-terminus.,Drs2p with a Thrombin cleavable C-terminal biotin acceptor domain (BAD) tag, and additional Thrombin cleavage site in the C-terminus. Number of copies: 1 / Enantiomer: LEVO / EC number: P-type phospholipid transporter |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 164.175359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNDDRETPPK RKPGEDDTLF DIDFLDDTTS HSGSRSKVTN SHANANYIPP SHVLPEETID LDADDDNIEN DVHENLFMSN NHDDQTSWN ANRFDSDAYQ PQSLRAVKPP GLFARFGNGL KNAFTFKRKK GPESFEMNHY NAVTNNELDD NYLDSRNKFN I KILFNRYI ...String: MNDDRETPPK RKPGEDDTLF DIDFLDDTTS HSGSRSKVTN SHANANYIPP SHVLPEETID LDADDDNIEN DVHENLFMSN NHDDQTSWN ANRFDSDAYQ PQSLRAVKPP GLFARFGNGL KNAFTFKRKK GPESFEMNHY NAVTNNELDD NYLDSRNKFN I KILFNRYI LRKNVGDAEG NGEPRVIHIN DSLANSSFGY SDNHISTTKY NFATFLPKFL FQEFSKYANL FFLCTSAIQQ VP HVSPTNR YTTIGTLLVV LIVSAMKECI EDIKRANSDK ELNNSTAEIF SEAHDDFVEK RWIDIRVGDI IRVKSEEPIP ADT IILSSS EPEGLCYIET ANLDGETNLK IKQSRVETAK FIDVKTLKNM NGKVVSEQPN SSLYTYEGTM TLNDRQIPLS PDQM ILRGA TLRNTAWIFG LVIFTGHETK LLRNATATPI KRTAVEKIIN RQIIALFTVL IVLILISSIG NVIMSTADAK HLSYL YLEG TNKAGLFFKD FLTFWILFSN LVPISLFVTV ELIKYYQAFM IGSDLDLYYE KTDTPTVVRT SSLVEELGQI EYIFSD KTG TLTRNIMEFK SCSIAGHCYI DKIPEDKTAT VEDGIEVGYR KFDDLKKKLN DPSDEDSPII NDFLTLLATC HTVIPEF QS DGSIKYQAAS PDEGALVQGG ADLGYKFIIR KPNSVTVLLE ETGEEKEYQL LNICEFNSTR KRMSAIFRFP DGSIKLFC K GADTVILERL DDEANQYVEA TMRHLEDYAS EGLRTLCLAM RDISEGEYEE WNSIYNEAAT TLDNRAEKLD EAANLIEKN LILIGATAIE DKLQDGVPET IHTLQEAGIK IWVLTGDRQE TAINIGMSCR LLSEDMNLLI INEETRDDTE RNLLEKINAL NEHQLSTHD MNTLALVIDG KSLGFALEPE LEDYLLTVAK LCKAVICCRV SPLQKALVVK MVKRKSSSLL LAIGDGANDV S MIQAAHVG VGISGMEGMQ AARSADIAVG QFKFLKKLLL VHGSWSYQRI SVAILYSFYK NTALYMTQFW YVFANAFSGQ SI MESWTMS FYNLFFTVWP PFVIGVFDQF VSSRLLERYP QLYKLGQKGQ FFSVYIFWGW IINGFFHSAI VFIGTILIYR YGF ALNMHG ELADHWSWGV TVYTTSVIIV LGKAALVTNQ WTKFTLIAIP GSLLFWLIFF PIYASIFPHA NISREYYGVV KHTY GSGVF WLTLIVLPIF ALVRDFLWKY YKRMYEPETY HVIQEMQKYN ISLVPRGSDS RPHVQQFQNA IRKVRQVQRM KKQRG FAFS QAEEGGQEKI VRMYDTTQKR GKYGELQDAS ANPFNDNNGL GSNDFESAEP FIENPFADGN QNSNRFSSSR DDISFD IGG GGLVPRGSGG TAAAPGPAPA PAPASAPAAA APAGAGTPVT APLAGTIWKV LASEGQTVAA GEVLLILEAM KMETEIR AA QAGTVRGIAV KAGDAVAVGD TLM UniProtKB: Phospholipid-transporting ATPase DRS2, Phospholipid-transporting ATPase DRS2 |
-Macromolecule #2: Cell division control protein 50
| Macromolecule | Name: Cell division control protein 50 / type: protein_or_peptide / ID: 2 / Details: Cdc50p with Thrombin cleavable C-terminal His-tag / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 47.371797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVSLFKRGKA PPLTKEGPTS KKPPNTAFRQ QRLKAWQPIL SPQSVLPLLI FVACIFTPIG IGLIVSATKV QDLTIDYSHC DTKASTTAF EDIPKKYIKY HFKSKVENKP QWRLTENENG EQSCELQFEI PNDIKKSIFI YYKITNFYQN HRRYVQSFDT K QILGEPIK ...String: MVSLFKRGKA PPLTKEGPTS KKPPNTAFRQ QRLKAWQPIL SPQSVLPLLI FVACIFTPIG IGLIVSATKV QDLTIDYSHC DTKASTTAF EDIPKKYIKY HFKSKVENKP QWRLTENENG EQSCELQFEI PNDIKKSIFI YYKITNFYQN HRRYVQSFDT K QILGEPIK KDDLDTSCSP IRSREDKIIY PCGLIANSMF NDTFSQVLSG IDDTEDYNLT NKHISWSIDR HRFKTTKYNA SD IVPPPNW MKKYPDGYTD ENLPDIHTWE EFQVWMRTAA FPKFYKLTLK NESASLPKGK YQMNIELNYP ISLFGGTKSF VLT TNGAIG GRNMSLGVLY LIVAGLCALF GIIFLVKLIF QPRAMGDHTY LNFDDEENED YEDVHAENTT LREILGGGGL VPRG SGGHH HHHHHHHH UniProtKB: Phospholipid-transporting ATPase accessory subunit CDC50 |
-Macromolecule #5: (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-...
| Macromolecule | Name: (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate type: ligand / ID: 5 / Number of copies: 1 / Formula: 2Y5 |
|---|---|
| Molecular weight | Theoretical: 967.108 Da |
| Chemical component information |  ChemComp-2Y5: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 3 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | Purified in detergent lauryl maltose neopentyl glycol (LMNG) |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 2074 / Average exposure time: 1.5 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Domains were rigid body fit into the density and manually adjusted and reconnected. |
| Refinement | Space: REAL / Overall B value: 55 / Target criteria: correlation coefficient |
| Output model |  PDB-7oh4: |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)