+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12878 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA-free Ribonuclease P from Halorhodospira halophila | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNAseP / metallonuclease / HARP / HYDROLASE | |||||||||
| Function / homology | RNA-free ribonuclease P / PINc domain ribonuclease / ribonuclease P / ribonuclease P activity / tRNA 5'-leader removal / PIN-like domain superfamily / RNA-free ribonuclease P Function and homology information Function and homology information | |||||||||
| Biological species |  Halorhodospira halophila SL1 (bacteria) / Halorhodospira halophila SL1 (bacteria) /  Halorhodospira halophila (strain DSM 244 / SL1) (bacteria) Halorhodospira halophila (strain DSM 244 / SL1) (bacteria) | |||||||||
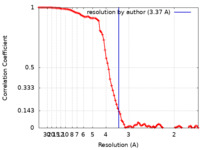
| Method | single particle reconstruction / cryo EM / Resolution: 3.37 Å | |||||||||
 Authors Authors | Altegoer F / Bange G | |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Structure and mechanistic features of the prokaryotic minimal RNase P. Authors: Rebecca Feyh / Nadine B Waeber / Simone Prinz / Pietro Ivan Giammarinaro / Gert Bange / Georg Hochberg / Roland K Hartmann / Florian Altegoer /  Abstract: Endonucleolytic removal of 5'-leader sequences from tRNA precursor transcripts (pre-tRNAs) by ribonuclease P (RNase P) is essential for protein synthesis. Beyond RNA-based RNase P enzymes, protein- ...Endonucleolytic removal of 5'-leader sequences from tRNA precursor transcripts (pre-tRNAs) by ribonuclease P (RNase P) is essential for protein synthesis. Beyond RNA-based RNase P enzymes, protein-only versions of the enzyme exert this function in various eukarya (there termed PRORPs) and in some bacteria ( and close relatives); both enzyme types belong to distinct subgroups of the PIN domain metallonuclease superfamily. Homologs of RNase P (HARPs) are also expressed in some other bacteria and many archaea, where they coexist with RNA-based RNase P and do not represent the main RNase P activity. Here, we solved the structure of the bacterial HARP from by cryo-electron microscopy, revealing a novel screw-like dodecameric assembly. Biochemical experiments demonstrate that oligomerization is required for RNase P activity of HARPs. We propose that the tRNA substrate binds to an extended spike-helix (SH) domain that protrudes from the screw-like assembly to position the 5'-end in close proximity to the active site of the neighboring dimer. The structure suggests that eukaryotic PRORPs and prokaryotic HARPs recognize the same structural elements of pre-tRNAs (tRNA elbow region and cleavage site). Our analysis thus delivers the structural and mechanistic basis for pre-tRNA processing by the prokaryotic HARP system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12878.map.gz emd_12878.map.gz | 33 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12878-v30.xml emd-12878-v30.xml emd-12878.xml emd-12878.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12878_fsc.xml emd_12878_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_12878.png emd_12878.png | 119.9 KB | ||
| Filedesc metadata |  emd-12878.cif.gz emd-12878.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12878 http://ftp.pdbj.org/pub/emdb/structures/EMD-12878 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12878 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12878 | HTTPS FTP |
-Validation report
| Summary document |  emd_12878_validation.pdf.gz emd_12878_validation.pdf.gz | 560.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12878_full_validation.pdf.gz emd_12878_full_validation.pdf.gz | 559.8 KB | Display | |
| Data in XML |  emd_12878_validation.xml.gz emd_12878_validation.xml.gz | 11 KB | Display | |
| Data in CIF |  emd_12878_validation.cif.gz emd_12878_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12878 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12878 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12878 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12878 | HTTPS FTP |
-Related structure data
| Related structure data |  7og5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12878.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12878.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.833 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Dodecameric assembly of minimal RNAseP system from Halorhodospira...
| Entire | Name: Dodecameric assembly of minimal RNAseP system from Halorhodospira halophila |
|---|---|
| Components |
|
-Supramolecule #1: Dodecameric assembly of minimal RNAseP system from Halorhodospira...
| Supramolecule | Name: Dodecameric assembly of minimal RNAseP system from Halorhodospira halophila type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Halorhodospira halophila SL1 (bacteria) Halorhodospira halophila SL1 (bacteria) |
| Molecular weight | Theoretical: 390 KDa |
-Macromolecule #1: RNA-free ribonuclease P
| Macromolecule | Name: RNA-free ribonuclease P / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO / EC number: ribonuclease P |
|---|---|
| Source (natural) | Organism:  Halorhodospira halophila (strain DSM 244 / SL1) (bacteria) Halorhodospira halophila (strain DSM 244 / SL1) (bacteria)Strain: DSM 244 / SL1 |
| Molecular weight | Theoretical: 24.051338 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMASRRFV LDTSVFTNPD VYLRFDEEPM QAISVFLGLA RRADAEFYMP GPVYQELCNL RSMDLIGAEF ETEVYIRSPR RFSMTIPSE VLYEFIEEVR TRIQRGLRIA EEHARQAGQA ESLPPELITQ LRERYREAMR RGILDSREDI DVVLLAYELD A TLVSADEG ...String: GSHMASRRFV LDTSVFTNPD VYLRFDEEPM QAISVFLGLA RRADAEFYMP GPVYQELCNL RSMDLIGAEF ETEVYIRSPR RFSMTIPSE VLYEFIEEVR TRIQRGLRIA EEHARQAGQA ESLPPELITQ LRERYREAMR RGILDSREDI DVVLLAYELD A TLVSADEG MRKFAERIGI KLVNPRYLRG VMQNLAGDDP GHAPPCGPDQ PAG UniProtKB: RNA-free ribonuclease P |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: Solutions were prepared freshly and filtered through a 0.2 um filter | |||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 11s with blot force -1 before plunging. | |||||||||
| Details | The sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 8393 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 181 |
|---|---|
| Output model |  PDB-7og5: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)