[English] 日本語
 Yorodumi
Yorodumi- EMDB-8511: Structure of the human HCN1 hyperpolarization-activated cyclic nu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8511 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human HCN1 hyperpolarization-activated cyclic nucleotide-gated ion channel | |||||||||
 Map data Map data | Human HCN1 hyperpolarization-activated cyclic nucleotide-gated ion channel, low-passed filtered to 3.5 A, B-factor sharpened at -90 A2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pacemaker ion channel / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information: / positive regulation of membrane hyperpolarization / HCN channels / general adaptation syndrome, behavioral process / HCN channel complex / retinal cone cell development / intracellularly cAMP-activated cation channel activity / negative regulation of action potential / regulation of SA node cell action potential / apical dendrite ...: / positive regulation of membrane hyperpolarization / HCN channels / general adaptation syndrome, behavioral process / HCN channel complex / retinal cone cell development / intracellularly cAMP-activated cation channel activity / negative regulation of action potential / regulation of SA node cell action potential / apical dendrite / maternal behavior / sodium ion import across plasma membrane / response to L-glutamate / apical protein localization / negative regulation of synaptic transmission, glutamatergic / voltage-gated monoatomic cation channel activity / voltage-gated sodium channel activity / regulation of membrane depolarization / potassium ion import across plasma membrane / phosphatidylinositol-3,4,5-trisphosphate binding / regulation of heart rate by cardiac conduction / voltage-gated potassium channel activity / potassium channel activity / cAMP binding / neuronal action potential / cellular response to interferon-beta / phosphatidylinositol-4,5-bisphosphate binding / presynaptic active zone membrane / potassium ion transmembrane transport / dendrite membrane / axon terminus / cellular response to cAMP / sodium ion transmembrane transport / dendritic shaft / regulation of membrane potential / response to calcium ion / basolateral plasma membrane / protein homotetramerization / postsynaptic membrane / axon / neuronal cell body / dendrite / glutamatergic synapse / cell surface / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Lee C-H / MacKinnon R | |||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: Structures of the Human HCN1 Hyperpolarization-Activated Channel. Authors: Chia-Hsueh Lee / Roderick MacKinnon /  Abstract: Hyperpolarization-activated cyclic nucleotide-gated (HCN) channels underlie the control of rhythmic activity in cardiac and neuronal pacemaker cells. In HCN, the polarity of voltage dependence is ...Hyperpolarization-activated cyclic nucleotide-gated (HCN) channels underlie the control of rhythmic activity in cardiac and neuronal pacemaker cells. In HCN, the polarity of voltage dependence is uniquely reversed. Intracellular cyclic adenosine monophosphate (cAMP) levels tune the voltage response, enabling sympathetic nerve stimulation to increase the heart rate. We present cryo-electron microscopy structures of the human HCN channel in the absence and presence of cAMP at 3.5 Å resolution. HCN channels contain a K channel selectivity filter-forming sequence from which the amino acids create a unique structure that explains Na and K permeability. The voltage sensor adopts a depolarized conformation, and the pore is closed. An S4 helix of unprecedented length extends into the cytoplasm, contacts the C-linker, and twists the inner helical gate shut. cAMP binding rotates cytoplasmic domains to favor opening of the inner helical gate. These structures advance understanding of ion selectivity, reversed polarity gating, and cAMP regulation in HCN channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8511.map.gz emd_8511.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8511-v30.xml emd-8511-v30.xml emd-8511.xml emd-8511.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8511.png emd_8511.png | 142.4 KB | ||
| Filedesc metadata |  emd-8511.cif.gz emd-8511.cif.gz | 5.3 KB | ||
| Others |  emd_8511_additional.map.gz emd_8511_additional.map.gz | 45.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8511 http://ftp.pdbj.org/pub/emdb/structures/EMD-8511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8511 | HTTPS FTP |
-Related structure data
| Related structure data |  5u6oMC  8512C  5u6pC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10081 (Title: Structure of the human HCN1 hyperpolarization-activated cyclic nucleotide-gated ion channel EMPIAR-10081 (Title: Structure of the human HCN1 hyperpolarization-activated cyclic nucleotide-gated ion channelData size: 66.5 Data #1: Aligned summed micrographs of the human HCN1 hyperpolarization-activated channel [micrographs - single frame] Data #2: Shiny particle image stacks of the human HCN1 hyperpolarization-activated channel [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8511.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8511.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human HCN1 hyperpolarization-activated cyclic nucleotide-gated ion channel, low-passed filtered to 3.5 A, B-factor sharpened at -90 A2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
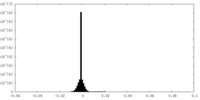
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
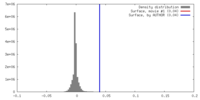
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Human HCN1 hyperpolarization-activated cyclic nucleotide-gated ion channel: unfiltered,...
| File | emd_8511_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human HCN1 hyperpolarization-activated cyclic nucleotide-gated ion channel: unfiltered, unsharpened map | ||||||||||||
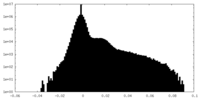
| Projections & Slices |
| ||||||||||||
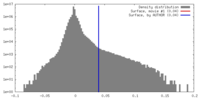
| Density Histograms |
- Sample components
Sample components
-Entire : Human HCN1 hyperpolarization-activated cyclic nucleotide-gated io...
| Entire | Name: Human HCN1 hyperpolarization-activated cyclic nucleotide-gated ion channel |
|---|---|
| Components |
|
-Supramolecule #1: Human HCN1 hyperpolarization-activated cyclic nucleotide-gated io...
| Supramolecule | Name: Human HCN1 hyperpolarization-activated cyclic nucleotide-gated ion channel type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Potassium/sodium hyperpolarization-activated cyclic nucleotide-ga...
| Macromolecule | Name: Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 74.643734 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEGGGKPNSS SNSRDDGNSV FPAKASATGA GPAAAEKRLG TPPGGGGAGA KEHGNSVCFK VDGGGGGGGG GGGGEEPAGG FEDAEGPRR QYGFMQRQFT SMLQPGVNKF SLRMFGSQKA VEKEQERVKT AGFWIIHPYS DFRFYWDLIM LIMMVGNLVI I PVGITFFT ...String: MEGGGKPNSS SNSRDDGNSV FPAKASATGA GPAAAEKRLG TPPGGGGAGA KEHGNSVCFK VDGGGGGGGG GGGGEEPAGG FEDAEGPRR QYGFMQRQFT SMLQPGVNKF SLRMFGSQKA VEKEQERVKT AGFWIIHPYS DFRFYWDLIM LIMMVGNLVI I PVGITFFT EQTTTPWIIF NVASDTVFLL DLIMNFRTGT VNEDSSEIIL DPKVIKMNYL KSWFVVDFIS SIPVDYIFLI VE KGMDSEV YKTARALRIV RFTKILSLLR LLRLSRLIRY IHQWEEIFHM TYDLASAVVR IFNLIGMMLL LCHWDGCLQF LVP LLQDFP PDCWVSLNEM VNDSWGKQYS YALFKAMSHM LCIGYGAQAP VSMSDLWITM LSMIVGATCY AMFVGHATAL IQSL DSSRR QYQEKYKQVE QYMSFHKLPA DMRQKIHDYY EHRYQGKIFD EENILNELND PLREEIVNFN CRKLVATMPL FANAD PNFV TAMLSKLRFE VFQPGDYIIR EGAVGKKMYF IQHGVAGVIT KSSKEMKLTD GSYFGEICLL TKGRRTASVR ADTYCR LYS LSVDNFNEVL EEYPMMRRAF ETVAIDRLDR IGKKNSILLQ KFQKDLNTGV FNNQENEILK QIVKHDREMV QAALPRE SS SVLNTDPDAE KPRFASNL UniProtKB: Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 1.26 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.3000000000000003 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C4 (4 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 55870 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)