[English] 日本語
 Yorodumi
Yorodumi- EMDB-12763: Human mitochondrial ribosome large subunit assembly intermediate ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12763 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human mitochondrial ribosome large subunit assembly intermediate with MTERF4-NSUN4, MRM2, MTG1, the MALSU module, GTPBP5 and mtEF-Tu | |||||||||
 Map data Map data | Locally filtered map from cryoSPARC non-uniform refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mitochondria / GTPase / Ribosome assembly intermediate / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationmRNA (cytidine-5-)-methyltransferase activity / rRNA modification in the mitochondrion / regulation of respiratory system process / mitochondrial RNA modification / mitochondrial RNA catabolic process / regulation of mitochondrial translation / negative regulation of mitochondrial translation / rRNA (uridine-2'-O-)-methyltransferase activity / rRNA 2'-O-methylation / rRNA (cytosine-C5-)-methyltransferase activity ...mRNA (cytidine-5-)-methyltransferase activity / rRNA modification in the mitochondrion / regulation of respiratory system process / mitochondrial RNA modification / mitochondrial RNA catabolic process / regulation of mitochondrial translation / negative regulation of mitochondrial translation / rRNA (uridine-2'-O-)-methyltransferase activity / rRNA 2'-O-methylation / rRNA (cytosine-C5-)-methyltransferase activity / SARS-CoV-2 modulates autophagy / mitochondrial large ribosomal subunit assembly / negative regulation of ribosome biogenesis / Mitochondrial Fatty Acid Beta-Oxidation / Protein lipoylation / positive regulation of mitochondrial translation / RNA methylation / Complex I biogenesis / RNA methyltransferase activity / Respiratory electron transport / protein lipoylation / rRNA methyltransferase activity / mitochondrial transcription / mitochondrial translational termination / Mitochondrial ribosome-associated quality control / mitochondrial translational elongation / mitochondrial ribosome assembly / Mitochondrial translation elongation / translation release factor activity, codon nonspecific / Mitochondrial translation initiation / Mitochondrial translation termination / camera-type eye development / mitochondrial fission / iron-sulfur cluster assembly complex / mitochondrial large ribosomal subunit binding / mitochondrial large ribosomal subunit / peptidyl-tRNA hydrolase / mitochondrial [2Fe-2S] assembly complex / translation release factor activity / mitochondrial ribosome / rRNA methylation / Hydrolases; Acting on ester bonds; Endoribonucleases producing 5'-phosphomonoesters / mitochondrial small ribosomal subunit / peptidyl-tRNA hydrolase activity / protein-synthesizing GTPase / [2Fe-2S] cluster assembly / mitochondrial translation / : / iron-sulfur cluster assembly / acyl binding / acyl carrier activity / ribosomal large subunit binding / proton motive force-driven mitochondrial ATP synthesis / mitochondrial nucleoid / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / translational elongation / respiratory chain complex I / anatomical structure morphogenesis / translation elongation factor activity / RNA processing / Mitochondrial protein degradation / rescue of stalled cytosolic ribosome / Transferases; Transferring one-carbon groups; Methyltransferases / fatty acid binding / aerobic respiration / cellular response to leukemia inhibitory factor / ribosomal large subunit biogenesis / methyltransferase activity / mitochondrial membrane / fibrillar center / fatty acid biosynthetic process / rRNA processing / cell junction / double-stranded RNA binding / large ribosomal subunit / heart development / double-stranded DNA binding / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / endonuclease activity / mitochondrial outer membrane / mitochondrial inner membrane / negative regulation of translation / rRNA binding / nuclear body / structural constituent of ribosome / ribosome / translation / mitochondrial matrix / ribonucleoprotein complex / protein domain specific binding / nucleotide binding / GTPase activity / mRNA binding / apoptotic process / calcium ion binding / regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / GTP binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
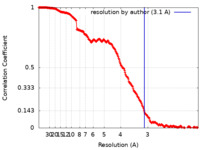
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Valentin Gese G / Hallberg BM | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis for late maturation steps of the human mitoribosomal large subunit. Authors: Miriam Cipullo / Genís Valentín Gesé / Anas Khawaja / B Martin Hällberg / Joanna Rorbach /   Abstract: Mitochondrial ribosomes (mitoribosomes) synthesize a critical set of proteins essential for oxidative phosphorylation. Therefore, mitoribosomal function is vital to the cellular energy supply. ...Mitochondrial ribosomes (mitoribosomes) synthesize a critical set of proteins essential for oxidative phosphorylation. Therefore, mitoribosomal function is vital to the cellular energy supply. Mitoribosome biogenesis follows distinct molecular pathways that remain poorly understood. Here, we determine the cryo-EM structures of mitoribosomes isolated from human cell lines with either depleted or overexpressed mitoribosome assembly factor GTPBP5, allowing us to capture consecutive steps during mitoribosomal large subunit (mt-LSU) biogenesis. Our structures provide essential insights into the last steps of 16S rRNA folding, methylation and peptidyl transferase centre (PTC) completion, which require the coordinated action of nine assembly factors. We show that mammalian-specific MTERF4 contributes to the folding of 16S rRNA, allowing 16 S rRNA methylation by MRM2, while GTPBP5 and NSUN4 promote fine-tuning rRNA rearrangements leading to PTC formation. Moreover, our data reveal an unexpected involvement of the elongation factor mtEF-Tu in mt-LSU assembly, where mtEF-Tu interacts with GTPBP5, similar to its interaction with tRNA during translational elongation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12763.map.gz emd_12763.map.gz | 52 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12763-v30.xml emd-12763-v30.xml emd-12763.xml emd-12763.xml | 92.6 KB 92.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12763_fsc.xml emd_12763_fsc.xml | 20.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_12763.png emd_12763.png | 61.4 KB | ||
| Filedesc metadata |  emd-12763.cif.gz emd-12763.cif.gz | 19.2 KB | ||
| Others |  emd_12763_additional_1.map.gz emd_12763_additional_1.map.gz emd_12763_half_map_1.map.gz emd_12763_half_map_1.map.gz emd_12763_half_map_2.map.gz emd_12763_half_map_2.map.gz | 47.9 MB 52.1 MB 52.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12763 http://ftp.pdbj.org/pub/emdb/structures/EMD-12763 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12763 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12763 | HTTPS FTP |
-Related structure data
| Related structure data |  7o9kMC  7o9mC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12763.map.gz / Format: CCP4 / Size: 56 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12763.map.gz / Format: CCP4 / Size: 56 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally filtered map from cryoSPARC non-uniform refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Map post-processed with DeepEMhancer
| File | emd_12763_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map post-processed with DeepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
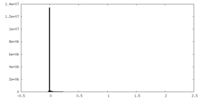
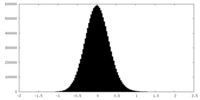
| Density Histograms |
-Half map: Half map A
| File | emd_12763_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
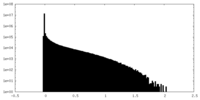
| Density Histograms |
-Half map: Half map B
| File | emd_12763_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 55S subunit assembly intermediate
+Supramolecule #1: 55S subunit assembly intermediate
+Macromolecule #1: 39S ribosomal protein L32, mitochondrial
+Macromolecule #2: 39S ribosomal protein L42, mitochondrial
+Macromolecule #3: 39S ribosomal protein L33, mitochondrial
+Macromolecule #4: 39S ribosomal protein L34, mitochondrial
+Macromolecule #5: 39S ribosomal protein L35, mitochondrial
+Macromolecule #6: 39S ribosomal protein L36, mitochondrial
+Macromolecule #7: 39S ribosomal protein L37, mitochondrial
+Macromolecule #8: 39S ribosomal protein L38, mitochondrial
+Macromolecule #9: 39S ribosomal protein L39, mitochondrial
+Macromolecule #10: 39S ribosomal protein L40, mitochondrial
+Macromolecule #11: 39S ribosomal protein L41, mitochondrial
+Macromolecule #13: 5-methylcytosine rRNA methyltransferase NSUN4
+Macromolecule #15: Transcription termination factor 4, mitochondrial
+Macromolecule #16: Mitochondrial ribosome-associated GTPase 1
+Macromolecule #17: 39S ribosomal protein L2, mitochondrial
+Macromolecule #18: 39S ribosomal protein L3, mitochondrial
+Macromolecule #19: 39S ribosomal protein L4, mitochondrial
+Macromolecule #20: 39S ribosomal protein L12, mitochondrial
+Macromolecule #21: Mitochondrial ribosome-associated GTPase 2
+Macromolecule #22: 39S ribosomal protein L9, mitochondrial
+Macromolecule #23: 39S ribosomal protein L10, mitochondrial
+Macromolecule #24: 39S ribosomal protein L11, mitochondrial
+Macromolecule #25: 39S ribosomal protein L13, mitochondrial
+Macromolecule #26: 39S ribosomal protein L14, mitochondrial
+Macromolecule #27: 39S ribosomal protein L15, mitochondrial
+Macromolecule #28: 39S ribosomal protein L16, mitochondrial
+Macromolecule #29: 39S ribosomal protein L17, mitochondrial
+Macromolecule #30: Mitochondrial ribosomal protein L18, isoform CRA_b
+Macromolecule #31: 39S ribosomal protein L19, mitochondrial
+Macromolecule #32: 39S ribosomal protein L20, mitochondrial
+Macromolecule #33: 39S ribosomal protein L21, mitochondrial
+Macromolecule #34: 39S ribosomal protein L22, mitochondrial
+Macromolecule #35: 39S ribosomal protein L23, mitochondrial
+Macromolecule #36: 39S ribosomal protein L24, mitochondrial
+Macromolecule #37: 39S ribosomal protein L27, mitochondrial
+Macromolecule #38: 39S ribosomal protein L28, mitochondrial
+Macromolecule #39: 39S ribosomal protein L47, mitochondrial
+Macromolecule #40: 39S ribosomal protein L30, mitochondrial
+Macromolecule #41: 39S ribosomal protein L43, mitochondrial
+Macromolecule #42: 39S ribosomal protein L44, mitochondrial
+Macromolecule #43: 39S ribosomal protein L45, mitochondrial
+Macromolecule #44: 39S ribosomal protein L46, mitochondrial
+Macromolecule #45: 39S ribosomal protein L48, mitochondrial
+Macromolecule #46: 39S ribosomal protein L49, mitochondrial
+Macromolecule #47: 39S ribosomal protein L50, mitochondrial
+Macromolecule #48: 39S ribosomal protein L51, mitochondrial
+Macromolecule #49: 39S ribosomal protein L52, mitochondrial
+Macromolecule #50: 39S ribosomal protein L53, mitochondrial
+Macromolecule #51: 39S ribosomal protein L54, mitochondrial
+Macromolecule #52: 39S ribosomal protein L55, mitochondrial
+Macromolecule #53: rRNA methyltransferase 2, mitochondrial
+Macromolecule #54: Ribosomal protein 63, mitochondrial
+Macromolecule #55: Peptidyl-tRNA hydrolase ICT1, mitochondrial
+Macromolecule #56: Growth arrest and DNA damage-inducible proteins-interacting protein 1
+Macromolecule #57: 39S ribosomal protein S18a, mitochondrial
+Macromolecule #58: 39S ribosomal protein S30, mitochondrial
+Macromolecule #59: Elongation factor Tu, mitochondrial
+Macromolecule #60: Mitochondrial assembly of ribosomal large subunit protein 1
+Macromolecule #61: MIEF1 upstream open reading frame protein
+Macromolecule #62: Acyl carrier protein, mitochondrial
+Macromolecule #63: UNK
+Macromolecule #12: 16S rRNA
+Macromolecule #14: MT-TRNAVAL
+Macromolecule #64: ZINC ION
+Macromolecule #65: MAGNESIUM ION
+Macromolecule #66: S-ADENOSYLMETHIONINE
+Macromolecule #67: GUANOSINE-5'-DIPHOSPHATE
+Macromolecule #68: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #69: S-ADENOSYL-L-HOMOCYSTEINE
+Macromolecule #70: 4'-PHOSPHOPANTETHEINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 49.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 20.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-7o9k: |
 Movie
Movie Controller
Controller







































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)