[English] 日本語
 Yorodumi
Yorodumi- EMDB-12672: Dimeric Photosystem I of a temperature sensitive mutant Chlamydom... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12672 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dimeric Photosystem I of a temperature sensitive mutant Chlamydomonas reinhardtii | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | chlamydomonas / photosystem I / temperature sensitive / water molecules / PHOTOSYNTHESIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationphotosynthesis, light harvesting in photosystem I / photosynthesis, light harvesting / chloroplast thylakoid lumen / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chlorophyll binding / chloroplast thylakoid membrane ...photosynthesis, light harvesting in photosystem I / photosynthesis, light harvesting / chloroplast thylakoid lumen / photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosystem II / chlorophyll binding / chloroplast thylakoid membrane / response to light stimulus / photosynthesis / 4 iron, 4 sulfur cluster binding / oxidoreductase activity / electron transfer activity / magnesium ion binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 17.1 Å | |||||||||
 Authors Authors | Caspy I / Nelson N | |||||||||
| Funding support |  Israel, 2 items Israel, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant. Authors: Ido Caspy / Tom Schwartz / Vinzenz Bayro-Kaiser / Mariia Fadeeva / Amit Kessel / Nir Ben-Tal / Nathan Nelson /  Abstract: Water molecules play a pivotal functional role in photosynthesis, primarily as the substrate for Photosystem II (PSII). However, their importance and contribution to Photosystem I (PSI) activity ...Water molecules play a pivotal functional role in photosynthesis, primarily as the substrate for Photosystem II (PSII). However, their importance and contribution to Photosystem I (PSI) activity remains obscure. Using a high-resolution cryogenic electron microscopy (cryo-EM) PSI structure from a Chlamydomonas reinhardtii temperature-sensitive photoautotrophic PSII mutant (TSP4), a conserved network of water molecules - dating back to cyanobacteria - was uncovered, mainly in the vicinity of the electron transport chain (ETC). The high-resolution structure illustrated that the water molecules served as a ligand in every chlorophyll that was missing a fifth magnesium coordination in the PSI core and in the light-harvesting complexes (LHC). The asymmetric distribution of the water molecules near the ETC branches modulated their electrostatic landscape, distinctly in the space between the quinones and FX. The data also disclosed the first observation of eukaryotic PSI oligomerisation through a low-resolution PSI dimer that was comprised of PSI-10LHC and PSI-8LHC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12672.map.gz emd_12672.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12672-v30.xml emd-12672-v30.xml emd-12672.xml emd-12672.xml | 31.3 KB 31.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12672_fsc.xml emd_12672_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_12672.png emd_12672.png | 79.9 KB | ||
| Filedesc metadata |  emd-12672.cif.gz emd-12672.cif.gz | 8.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12672 http://ftp.pdbj.org/pub/emdb/structures/EMD-12672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12672 | HTTPS FTP |
-Related structure data
| Related structure data |  7o01MC  7bgiC  7blxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12672.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12672.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.481 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
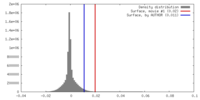
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Photosystem I of temperature sensitive Chlamydomonas reinhardtii
+Supramolecule #1: Photosystem I of temperature sensitive Chlamydomonas reinhardtii
+Macromolecule #1: Photosystem I P700 chlorophyll a apoprotein A1
+Macromolecule #2: Photosystem I P700 chlorophyll a apoprotein A2
+Macromolecule #3: Photosystem I iron-sulfur center
+Macromolecule #4: Photosystem I reaction center subunit II, chloroplastic
+Macromolecule #5: Photosystem I reaction center subunit IV, chloroplastic
+Macromolecule #6: Photosystem I reaction center subunit III, chloroplastic
+Macromolecule #7: Photosystem I reaction center subunit V, chloroplastic
+Macromolecule #8: Photosystem I reaction center subunit VIII
+Macromolecule #9: Photosystem I reaction center subunit IX
+Macromolecule #10: Photosystem I reaction center subunit psaK, chloroplastic
+Macromolecule #11: PSI subunit V
+Macromolecule #12: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #13: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #14: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #15: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #16: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #17: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #18: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #19: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #20: Chlorophyll a-b binding protein, chloroplastic
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 12418 / Average electron dose: 46.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 165000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)