登録情報 データベース : EMDB / ID : EMD-12667タイトル RNA polymerase II-Spt4/5-nucleosome-Chd1 structure - Map B Map B 複合体 : RNA polymerase II-Spt4/5-Chd1-nucleosome機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Saccharomyces cerevisiae (パン酵母)手法 / / 解像度 : 2.7 Å Farnung L / Ochmann M / Engeholm M / Cramer P 資金援助 European Union, Organization Grant number 国 European Research Council (ERC) 693023 European Union German Research Foundation (DFG) SFB860 German Research Foundation (DFG) SPP2191 German Research Foundation (DFG) EXC 2067/1- 390729940
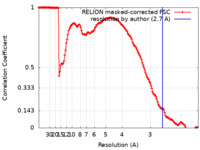
ジャーナル : Nat Struct Mol Biol / 年 : 2021タイトル : Structural basis of nucleosome transcription mediated by Chd1 and FACT.著者 : Lucas Farnung / Moritz Ochmann / Maik Engeholm / Patrick Cramer / 要旨 : Efficient transcription of RNA polymerase II (Pol II) through nucleosomes requires the help of various factors. Here we show biochemically that Pol II transcription through a nucleosome is ... Efficient transcription of RNA polymerase II (Pol II) through nucleosomes requires the help of various factors. Here we show biochemically that Pol II transcription through a nucleosome is facilitated by the chromatin remodeler Chd1 and the histone chaperone FACT when the elongation factors Spt4/5 and TFIIS are present. We report cryo-EM structures of transcribing Saccharomyces cerevisiae Pol II-Spt4/5-nucleosome complexes with bound Chd1 or FACT. In the first structure, Pol II transcription exposes the proximal histone H2A-H2B dimer that is bound by Spt5. Pol II has also released the inhibitory DNA-binding region of Chd1 that is poised to pump DNA toward Pol II. In the second structure, Pol II has generated a partially unraveled nucleosome that binds FACT, which excludes Chd1 and Spt5. These results suggest that Pol II progression through a nucleosome activates Chd1, enables FACT binding and eventually triggers transfer of FACT together with histones to upstream DNA. 履歴 登録 2021年3月24日 - ヘッダ(付随情報) 公開 2021年4月28日 - マップ公開 2021年4月28日 - 更新 2021年4月28日 - 現状 2021年4月28日 処理サイト : PDBe / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 機能・相同性情報
機能・相同性情報
 データ登録者
データ登録者 ドイツ, 4件
ドイツ, 4件  引用
引用 ジャーナル: Nat Struct Mol Biol / 年: 2021
ジャーナル: Nat Struct Mol Biol / 年: 2021
 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_12667.map.gz
emd_12667.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-12667-v30.xml
emd-12667-v30.xml emd-12667.xml
emd-12667.xml EMDBヘッダ
EMDBヘッダ emd_12667_fsc.xml
emd_12667_fsc.xml FSCデータファイル
FSCデータファイル emd_12667.png
emd_12667.png emd_12667_additional_1.map.gz
emd_12667_additional_1.map.gz emd_12667_half_map_1.map.gz
emd_12667_half_map_1.map.gz emd_12667_half_map_2.map.gz
emd_12667_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-12667
http://ftp.pdbj.org/pub/emdb/structures/EMD-12667 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12667
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12667 emd_12667_validation.pdf.gz
emd_12667_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_12667_full_validation.pdf.gz
emd_12667_full_validation.pdf.gz emd_12667_validation.xml.gz
emd_12667_validation.xml.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12667
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12667 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12667
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12667 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_12667.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_12667.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素

 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 ムービー
ムービー コントローラー
コントローラー

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)