[English] 日本語
 Yorodumi
Yorodumi- EMDB-1250: Structure of the E. coli signal recognition particle bound to a t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1250 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the E. coli signal recognition particle bound to a translating ribosome. | |||||||||
 Map data Map data | This is the map file of the E. coli signal recognition particle bound to a ribosome nascent chain complex. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsignal recognition particle / signal-recognition-particle GTPase / 7S RNA binding / SRP-dependent cotranslational protein targeting to membrane / GTPase activity / GTP binding / ATP hydrolysis activity Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 15.5 Å | |||||||||
 Authors Authors | Schaffitzel C / Oswald M / Berger I / Ishikawa T / Abrahams JP / Koerten HK / Koning RI / Ban N | |||||||||
 Citation Citation |  Journal: Nature / Year: 2006 Journal: Nature / Year: 2006Title: Structure of the E. coli signal recognition particle bound to a translating ribosome. Authors: Christiane Schaffitzel / Miro Oswald / Imre Berger / Takashi Ishikawa / Jan Pieter Abrahams / Henk K Koerten / Roman I Koning / Nenad Ban /  Abstract: The prokaryotic signal recognition particle (SRP) targets membrane proteins into the inner membrane. It binds translating ribosomes and screens the emerging nascent chain for a hydrophobic signal ...The prokaryotic signal recognition particle (SRP) targets membrane proteins into the inner membrane. It binds translating ribosomes and screens the emerging nascent chain for a hydrophobic signal sequence, such as the transmembrane helix of inner membrane proteins. If such a sequence emerges, the SRP binds tightly, allowing the SRP receptor to lock on. This assembly delivers the ribosome-nascent chain complex to the protein translocation machinery in the membrane. Using cryo-electron microscopy and single-particle reconstruction, we obtained a 16 A structure of the Escherichia coli SRP in complex with a translating E. coli ribosome containing a nascent chain with a transmembrane helix anchor. We also obtained structural information on the SRP bound to an empty E. coli ribosome. The latter might share characteristics with a scanning SRP complex, whereas the former represents the next step: the targeting complex ready for receptor binding. High-resolution structures of the bacterial ribosome and of the bacterial SRP components are available, and their fitting explains our electron microscopic density. The structures reveal the regions that are involved in complex formation, provide insight into the conformation of the SRP on the ribosome and indicate the conformational changes that accompany high-affinity SRP binding to ribosome nascent chain complexes upon recognition of the signal sequence. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1250.map.gz emd_1250.map.gz | 4.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1250-v30.xml emd-1250-v30.xml emd-1250.xml emd-1250.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  1250.gif 1250.gif | 39.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1250 http://ftp.pdbj.org/pub/emdb/structures/EMD-1250 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1250 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1250 | HTTPS FTP |
-Validation report
| Summary document |  emd_1250_validation.pdf.gz emd_1250_validation.pdf.gz | 230 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1250_full_validation.pdf.gz emd_1250_full_validation.pdf.gz | 229.1 KB | Display | |
| Data in XML |  emd_1250_validation.xml.gz emd_1250_validation.xml.gz | 5.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1250 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1250 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1250 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1250 | HTTPS FTP |
-Related structure data
| Related structure data |  2iy3MC  1251C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1250.map.gz / Format: CCP4 / Size: 5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1250.map.gz / Format: CCP4 / Size: 5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the map file of the E. coli signal recognition particle bound to a ribosome nascent chain complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.81 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
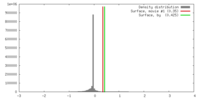
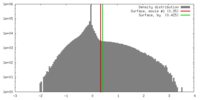
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : E. coli ribosome nascent chain complexes and signal recognition p...
| Entire | Name: E. coli ribosome nascent chain complexes and signal recognition particle |
|---|---|
| Components |
|
-Supramolecule #1000: E. coli ribosome nascent chain complexes and signal recognition p...
| Supramolecule | Name: E. coli ribosome nascent chain complexes and signal recognition particle type: sample / ID: 1000 / Oligomeric state: One SRP binds to one ribosome. / Number unique components: 5 |
|---|
-Supramolecule #1: E.coli 70S ribosome
| Supramolecule | Name: E.coli 70S ribosome / type: complex / ID: 1 / Recombinant expression: No / Ribosome-details: ribosome-prokaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.5 MDa |
-Macromolecule #1: A-site tRNA
| Macromolecule | Name: A-site tRNA / type: rna / ID: 1 / Classification: TRANSFER / Structure: DOUBLE HELIX / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25 KDa |
-Macromolecule #2: P-site tRNA
| Macromolecule | Name: P-site tRNA / type: rna / ID: 2 / Classification: TRANSFER / Structure: DOUBLE HELIX / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25 KDa |
-Macromolecule #3: E-site tRNA
| Macromolecule | Name: E-site tRNA / type: rna / ID: 3 / Classification: TRANSFER / Structure: DOUBLE HELIX / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25 KDa |
-Macromolecule #4: E. coli SRP
| Macromolecule | Name: E. coli SRP / type: ligand / ID: 4 / Name.synonym: Signal recognition particle / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 90 KDa |
| Recombinant expression | Organism: BL21StarDE3 / Recombinant plasmid: pET24a_Ffh and pUC19_Ffs |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 50 mM Hepes-KOH pH 7.5, 100 mM KCl, 25 mM MgCl2, 1 mM DTT, 1 mM GTP |
| Grid | Details: carbon-coated lacey formvar grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: home-built environmental chamber and vitrification device Method: blot for 1.5 sec before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Number real images: 251 / Average electron dose: 10 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 51000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: cryo stage / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 15.5 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Spider / Number images used: 24382 |
|---|
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: NOMAD |
| Details | Protocol: normal mode based flexible fitting. Fitting of SRP atomic model into isolated EM density |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: correlation coefficient |
| Output model |  PDB-2iy3: |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B |
|---|---|
| Software | Name: NOMAD |
| Details | Protocol: normal mode based flexible fitting. Fitting of SRP atomic model into isolated EM density |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: correlation coefficient |
| Output model |  PDB-2iy3: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)