+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12319 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
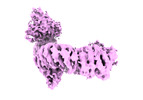
| Title | Nematocida Huwe1 in open conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Huwe1 / Nematocida / E3 Ligase / HECT / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationHECT-type E3 ubiquitin transferase / ubiquitin protein ligase activity / ubiquitin-dependent protein catabolic process / protein ubiquitination / membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Nematocida sp. ERTm5 (fungus) Nematocida sp. ERTm5 (fungus) | |||||||||
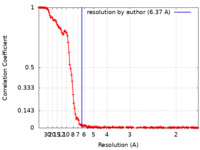
| Method | single particle reconstruction / cryo EM / Resolution: 6.37 Å | |||||||||
 Authors Authors | Petrova O / Grishkovskaya I | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Crystal structure of HUWE1: One ring to ubiquitinate them all Authors: Grabarrczyk DB / Petrova OA / Haselbach D / Kessler D / Clausen T | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12319.map.gz emd_12319.map.gz | 228 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12319-v30.xml emd-12319-v30.xml emd-12319.xml emd-12319.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12319_fsc.xml emd_12319_fsc.xml | 18.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_12319.png emd_12319.png | 108.9 KB | ||
| Masks |  emd_12319_msk_1.map emd_12319_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12319.cif.gz emd-12319.cif.gz | 8 KB | ||
| Others |  emd_12319_half_map_1.map.gz emd_12319_half_map_1.map.gz emd_12319_half_map_2.map.gz emd_12319_half_map_2.map.gz | 194.6 MB 194.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12319 http://ftp.pdbj.org/pub/emdb/structures/EMD-12319 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12319 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12319 | HTTPS FTP |
-Validation report
| Summary document |  emd_12319_validation.pdf.gz emd_12319_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12319_full_validation.pdf.gz emd_12319_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_12319_validation.xml.gz emd_12319_validation.xml.gz | 21.1 KB | Display | |
| Data in CIF |  emd_12319_validation.cif.gz emd_12319_validation.cif.gz | 27.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12319 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12319 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12319 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12319 | HTTPS FTP |
-Related structure data
| Related structure data |  7nh3MC  7nh1C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12319.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12319.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
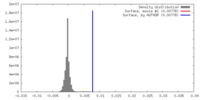
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12319_msk_1.map emd_12319_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
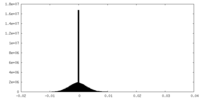
| Density Histograms |
-Half map: #1
| File | emd_12319_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12319_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Huwe1
| Entire | Name: Huwe1 |
|---|---|
| Components |
|
-Supramolecule #1: Huwe1
| Supramolecule | Name: Huwe1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Nematocida sp. ERTm5 (fungus) Nematocida sp. ERTm5 (fungus) |
| Molecular weight | Theoretical: 287 KDa |
-Macromolecule #1: E3 ubiquitin-protein ligase HUWE1
| Macromolecule | Name: E3 ubiquitin-protein ligase HUWE1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nematocida sp. ERTm5 (fungus) Nematocida sp. ERTm5 (fungus) |
| Molecular weight | Theoretical: 287.751281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKIERPMERR FSQPGKQISN TIKQLSNLPE EELPTYLNNI YEWRCAKTDL LYWVDVLDRF DEILKAVVTE YGLGQFQNKP MKESDKEMV YAILKFQKLL VENSSNKSMF SSFDVVEPFI YSFELDLAIE ALYLVSFFAS KIHIQRSIKT SMALMKMETL K TLIERVKE ...String: MKIERPMERR FSQPGKQISN TIKQLSNLPE EELPTYLNNI YEWRCAKTDL LYWVDVLDRF DEILKAVVTE YGLGQFQNKP MKESDKEMV YAILKFQKLL VENSSNKSMF SSFDVVEPFI YSFELDLAIE ALYLVSFFAS KIHIQRSIKT SMALMKMETL K TLIERVKE KPQSMYTYYD ETTNNCKRVS IKKAVKKGIY SISDKMLPSQ EIRRFIHAIR LHEMAEQHEK LKIIRMLAFS AL VYYNYTD LPIDSEFISR DLPETLAIIN TESYQMREAA VTMIDAIFRM RIRHSAVIAA MNAQSHEGMI MNLLKKVVNE DVP EHFAIV FFNFLSSCFA SGPCVSALFS AGIVQYVCNT LRDQPDISYR KRMRLVMGAN TFLFTLPAPF TWFISENGIR ILSK ELLLA VDVALGNIKD HDLLMYINSI MKIISQLFKN AGTAEAMRGF LEGEFPRAIS LILFHSEEFT PSILAYLFSA VGDYI HNEP SNLPFIIEAG IFDGFVMCMK KELPESPDFL LELPNLIEAF FLNSELIKKI DEENILDRIF ESFEIVNLSN IILMYD IGR AYGIFLENLV RHYPIISLTV KEKIYNTMKN LESMIPNTDP ELMRLLLGNL FRMMHRAVYR KTTNNQLITD KGLLNNR II SMLMLIEIPT ETELYTDVMN ILMEVFDEDQ TYVISYIAKV IDRVMKNKVT LENIEKIKRI LLIVNYLIFK NEETCEEF I KHCTSKGFLQ IVKRISQYFD NLKKDTALVS VAKNESLTNL YYSFTMGILK TVYRYSKTNA KEYLKIFGLL VEDLLSKTE QIDHLYYTQR MHGLKNYIMV DKSPQQYPQK NEFVVTQEYL MGINLSETLM EHAKNSVSML KDEKITMDAH KKVVSSILEV LSIYVRGIK NLFVPVKEPK KKVCEIEEHV LIIVELLMDY IDKSSIEHLL NIVKTAFFVT LRKQITKEMD RENEGSAEYK R RNILGSFF ISVFLNLPKE ETEAVLKSDI FTILVKNKKT FKQVYNKNCF GLLQYAAIKE SRVFSHLVSF IPKMTKQVGA ST DPAFLKL YAVGLVFVAM RDTLKKTTVL SRFIGNIVFA ESTCPAVVEA QGIVILAIVK LKIEYNLAIT IEDPIIHLNR IYA IVETAE QKEGLVILAN LLIRRVIESK EAAKEAVDTI IKNKHLGKSR MYTVHTVCSN LRNALFYSIS SFLEYLSSNF ECIS AGWEE IRHKLPDESA NPTESGQISE AVLETLSDGL FTKQIKIYSG LNAELFRTLF VHRATGKMEQ IVRIHSLCLL VVSFP QLIS TLVPEDYSFF MYFLENHAAY SKSLKPSAFQ QEDKTLAYWS GHFIMLIFNH TTYVEVKKYI LEKVLELLPT SINATV IFS ELIQEILTMR FSKNTFDENT ALVKEMNCLE VMIRSTMQID NRRKDYGSIM EILTKPMEYI TRILAVDEKE AFYEEVT QS EEEVYFEDID EGYMEDFDTD ETMDSDQVVY DDTEENSQEV GELCSEDSLT VYTAESNNYG EYLMSDEESS SEEEGMDE E QSSSEQSEKN EFLKSLCILP ISKLIGKEME IFNADERMPF VQKILEGSII SNLQEKEPSS TDEETFDSEE GSYRRHQYL MRREEENDDE ELNDYAIDPT DPDAHLDDEG PEEIDEGVDE YDDESQYDYD GNGSDDEEYD DEESFATGEE IAIGDENGEI PELDVEVLN NLPSSILEDT VENFYQDRIS SSTEYRAISL HFLNRLREEV RSVFEEHEAR YMETFAGEIV PRREEKKKKP Q EMPFIAER EAAASVPIDI VFGLIHMVLQ CGNRRNLYRI IHNISANKEV RIFSVETLVN SIYQAVVEGA SSSAGAGSST VN ASAGSSG NNEVIAPEVI TKRGFEALTY LCTKSSDFTT VFSYNTELIN KILQATNKRT ISESVKLLST VGDCFNNDKV AEP ENIEVR KYIGFLEYDM TDDTFKHFCE FIKKTDRFYR PMYLLYLMGG SKKSLEECLD KKAEFNSHTH HKGIIKLVRM LSLV NIMGI TETYLDSLME LREMPFWEYY FNIILPKEKE SLYASSILPL FKAFVIVHTI QMYIGRNENI NEFSEIPNSD SSIYY SVVE KEKDLINTFI QADPDLLFHA FAGLQKKILD FDNKRIYFYK KIREDVQLRP TISLMVQRGA VFEDTFHQLM RLNGEQ VRN AKFNIKFAGE EGVDAGGLTR EWYSELSKEM FNANYALFTP IGSSYQPNHI SHINPEHLVY FKFIGRIIGK AVYDEMT VD CHFTRAFYKR VLSIPVDLTD VEALDPEFHR SLVWILENDI ENVLDMTFSI EQDRFGITEI IDLKENGRNI AVTNENKR E YVELVCRFKL VRVIERQLSA FAEGFFEILD VDMLKMFNEK ELELLISGLP EIDVDDWRNN TIYFGYTSDS QVIRWYWRA VRNFSMEERA KLLQFATGTS KLPLEGFAGL RCQNGNQKFQ IHKASGGSSR LPTAHTCFNQ LDLPEYDSYE QLVKALLFSL EECTSGFGF A UniProtKB: HECT-type E3 ubiquitin transferase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 75 % / Chamber temperature: 4 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)