登録情報 データベース : EMDB / ID : EMD-12299タイトル Cryo-EM structure of NHEJ super-complex (dimer) NHEJ super-complex (dimer) anistropically sharpened map 細胞器官・細胞要素 : NHEJ super-complex (dimer)細胞器官・細胞要素 : DNA-dependent protein kinase catalytic subunitタンパク質・ペプチド : DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs細胞器官・細胞要素 : X-ray repair cross-complementing proteins 5 and 6タンパク質・ペプチド : X-ray repair cross-complementing protein 6タンパク質・ペプチド : X-ray repair cross-complementing protein 5細胞器官・細胞要素 : Repair protein and ligaseタンパク質・ペプチド : DNA repair protein XRCC4タンパク質・ペプチド : DNA ligase 4タンパク質・ペプチド : Non-homologous end-joining factor 1細胞器官・細胞要素 : DNADNA : DNA (27-MER)DNA : DNA (28-MER)DNA : DNA (27-MER) / / / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
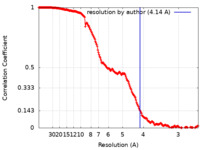
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト)手法 / / 解像度 : 4.14 Å Chaplin AK / Hardwick SW 資金援助 Organization Grant number 国 Wellcome Trust 200814/Z/16/Z; 2016
ジャーナル : Mol Cell / 年 : 2021タイトル : Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.著者: Amanda K Chaplin / Steven W Hardwick / Antonia Kefala Stavridi / Christopher J Buehl / Noah J Goff / Virginie Ropars / Shikang Liang / Taiana Maia De Oliveira / Dimitri Y Chirgadze / Katheryn ... 著者 : Amanda K Chaplin / Steven W Hardwick / Antonia Kefala Stavridi / Christopher J Buehl / Noah J Goff / Virginie Ropars / Shikang Liang / Taiana Maia De Oliveira / Dimitri Y Chirgadze / Katheryn Meek / Jean-Baptiste Charbonnier / Tom L Blundell / 要旨 : Non-homologous end joining (NHEJ) is one of two critical mechanisms utilized in humans to repair DNA double-strand breaks (DSBs). Unrepaired or incorrect repair of DSBs can lead to apoptosis or ... Non-homologous end joining (NHEJ) is one of two critical mechanisms utilized in humans to repair DNA double-strand breaks (DSBs). Unrepaired or incorrect repair of DSBs can lead to apoptosis or cancer. NHEJ involves several proteins, including the Ku70/80 heterodimer, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), X-ray cross-complementing protein 4 (XRCC4), XRCC4-like factor (XLF), and ligase IV. These core proteins bind DSBs and ligate the damaged DNA ends. However, details of the structural assembly of these proteins remain unclear. Here, we present cryo-EM structures of NHEJ supercomplexes that are composed of these core proteins and DNA, revealing the detailed structural architecture of this assembly. We describe monomeric and dimeric forms of this supercomplex and also propose the existence of alternate dimeric forms of long-range synaptic complexes. Finally, we show that mutational disruption of several structural features within these NHEJ complexes negatively affects DNA repair. 履歴 登録 2021年2月5日 - ヘッダ(付随情報) 公開 2021年8月18日 - マップ公開 2021年8月18日 - 更新 2024年7月10日 - 現状 2024年7月10日 処理サイト : PDBe / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) データ登録者
データ登録者 英国, 1件
英国, 1件  引用
引用 ジャーナル: Mol Cell / 年: 2021
ジャーナル: Mol Cell / 年: 2021


 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_12299.map.gz
emd_12299.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-12299-v30.xml
emd-12299-v30.xml emd-12299.xml
emd-12299.xml EMDBヘッダ
EMDBヘッダ emd_12299_fsc.xml
emd_12299_fsc.xml FSCデータファイル
FSCデータファイル emd_12299.png
emd_12299.png emd-12299.cif.gz
emd-12299.cif.gz emd_12299_half_map_1.map.gz
emd_12299_half_map_1.map.gz emd_12299_half_map_2.map.gz
emd_12299_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-12299
http://ftp.pdbj.org/pub/emdb/structures/EMD-12299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12299
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12299 emd_12299_validation.pdf.gz
emd_12299_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_12299_full_validation.pdf.gz
emd_12299_full_validation.pdf.gz emd_12299_validation.xml.gz
emd_12299_validation.xml.gz emd_12299_validation.cif.gz
emd_12299_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12299
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12299 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12299
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12299 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_12299.map.gz / 形式: CCP4 / 大きさ: 600.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_12299.map.gz / 形式: CCP4 / 大きさ: 600.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 ムービー
ムービー コントローラー
コントローラー





























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)