登録情報 データベース : EMDB / ID : EMD-12040タイトル Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-SKP1-SKP2-CKSHS1-p27~Ub~ARIH1. Transition State 2 複合体 : CUL1-RBX1-SKP1-SKP2-CKSHS1-p27~Ub~ARIH1複合体 : Cullin-1, E3 ubiquitin-protein ligase RBX1タンパク質・ペプチド : Cullin-1タンパク質・ペプチド : E3 ubiquitin-protein ligase RBX1複合体 : ARIH1, SKP2, CKS1B, SKP1, UBCタンパク質・ペプチド : E3 ubiquitin-protein ligase ARIH1タンパク質・ペプチド : S-phase kinase-associated protein 2タンパク質・ペプチド : Cyclin-dependent kinases regulatory subunit 1タンパク質・ペプチド : S-phase kinase-associated protein 1タンパク質・ペプチド : Polyubiquitin-C複合体 : Cyclin-dependent kinase inhibitor 1Bタンパク質・ペプチド : Cyclin-dependent kinase inhibitor 1Bリガンド : ZINC ION / / / / / / / / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
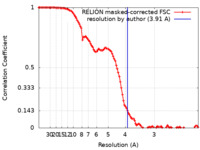
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト)手法 / / 解像度 : 3.91 Å Horn-Ghetko D / Prabu JR / Schulman BA 資金援助 Organization Grant number 国 European Research Council (ERC) H2020 789016-NEDD8Activate German Research Foundation (DFG) SCHU 3196/1-1
ジャーナル : Nature / 年 : 2021タイトル : Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.著者 : Daniel Horn-Ghetko / David T Krist / J Rajan Prabu / Kheewoong Baek / Monique P C Mulder / Maren Klügel / Daniel C Scott / Huib Ovaa / Gary Kleiger / Brenda A Schulman / 要旨 : E3 ligases are typically classified by hallmark domains such as RING and RBR, which are thought to specify unique catalytic mechanisms of ubiquitin transfer to recruited substrates. However, rather ... E3 ligases are typically classified by hallmark domains such as RING and RBR, which are thought to specify unique catalytic mechanisms of ubiquitin transfer to recruited substrates. However, rather than functioning individually, many neddylated cullin-RING E3 ligases (CRLs) and RBR-type E3 ligases in the ARIH family-which together account for nearly half of all ubiquitin ligases in humans-form E3-E3 super-assemblies. Here, by studying CRLs in the SKP1-CUL1-F-box (SCF) family, we show how neddylated SCF ligases and ARIH1 (an RBR-type E3 ligase) co-evolved to ubiquitylate diverse substrates presented on various F-box proteins. We developed activity-based chemical probes that enabled cryo-electron microscopy visualization of steps in E3-E3 ubiquitylation, initiating with ubiquitin linked to the E2 enzyme UBE2L3, then transferred to the catalytic cysteine of ARIH1, and culminating in ubiquitin linkage to a substrate bound to the SCF E3 ligase. The E3-E3 mechanism places the ubiquitin-linked active site of ARIH1 adjacent to substrates bound to F-box proteins (for example, substrates with folded structures or limited length) that are incompatible with previously described conventional RING E3-only mechanisms. The versatile E3-E3 super-assembly may therefore underlie widespread ubiquitylation. 履歴 登録 2020年12月5日 - ヘッダ(付随情報) 公開 2021年2月17日 - マップ公開 2021年2月17日 - 更新 2025年7月9日 - 現状 2025年7月9日 処理サイト : PDBe / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) データ登録者
データ登録者 ドイツ, 2件
ドイツ, 2件  引用
引用 ジャーナル: Nature / 年: 2021
ジャーナル: Nature / 年: 2021


 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_12040.map.gz
emd_12040.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-12040-v30.xml
emd-12040-v30.xml emd-12040.xml
emd-12040.xml EMDBヘッダ
EMDBヘッダ emd_12040_fsc.xml
emd_12040_fsc.xml FSCデータファイル
FSCデータファイル emd_12040.png
emd_12040.png emd_12040_msk_1.map
emd_12040_msk_1.map マスクマップ
マスクマップ emd-12040.cif.gz
emd-12040.cif.gz emd_12040_half_map_1.map.gz
emd_12040_half_map_1.map.gz emd_12040_half_map_2.map.gz
emd_12040_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-12040
http://ftp.pdbj.org/pub/emdb/structures/EMD-12040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12040
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12040 emd_12040_validation.pdf.gz
emd_12040_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_12040_full_validation.pdf.gz
emd_12040_full_validation.pdf.gz emd_12040_validation.xml.gz
emd_12040_validation.xml.gz emd_12040_validation.cif.gz
emd_12040_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12040
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12040 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12040
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12040 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_12040.map.gz / 形式: CCP4 / 大きさ: 115.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_12040.map.gz / 形式: CCP4 / 大きさ: 115.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) emd_12040_msk_1.map
emd_12040_msk_1.map 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 ムービー
ムービー コントローラー
コントローラー






















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






































 Trichoplusia ni (イラクサキンウワバ)
Trichoplusia ni (イラクサキンウワバ)