[English] 日本語
 Yorodumi
Yorodumi- EMDB-11820: Nonameric cytoplasmic domain of SctV from Yersinia enterocolitica -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11820 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
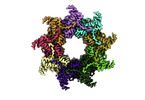
| Title | Nonameric cytoplasmic domain of SctV from Yersinia enterocolitica | ||||||||||||
 Map data Map data | Structure of the SctV cytoplasmic domain | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | T3SS / export apparatus / secretion / protein export / PROTEIN TRANSPORT | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) | ||||||||||||
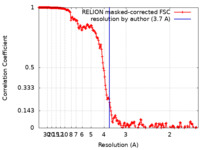
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Kuhlen L / Johnson S | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: PLoS One / Year: 2021 Journal: PLoS One / Year: 2021Title: Nonameric structures of the cytoplasmic domain of FlhA and SctV in the context of the full-length protein. Authors: Lucas Kuhlen / Steven Johnson / Jerry Cao / Justin C Deme / Susan M Lea /   Abstract: Type three secretion is the mechanism of protein secretion found in bacterial flagella and injectisomes. At its centre is the export apparatus (EA), a complex of five membrane proteins through which ...Type three secretion is the mechanism of protein secretion found in bacterial flagella and injectisomes. At its centre is the export apparatus (EA), a complex of five membrane proteins through which secretion substrates pass the inner membrane. While the complex formed by four of the EA proteins has been well characterised structurally, little is known about the structure of the membrane domain of the largest subunit, FlhA in flagella, SctV in injectisomes. Furthermore, the biologically relevant nonameric assembly of FlhA/SctV has been infrequently observed and differences in conformation of the cytoplasmic portion of FlhA/SctV between open and closed states have been suggested to reflect secretion system specific differences. FlhA has been shown to bind to chaperone-substrate complexes in an open state, but in previous assembled ring structures, SctV is in a closed state. Here, we identify FlhA and SctV homologues that can be recombinantly produced in the oligomeric state and study them using cryo-electron microscopy. The structures of the cytoplasmic domains from both FlhA and SctV are in the open state and we observe a conserved interaction between a short stretch of residues at the N-terminus of the cytoplasmic domain, known as FlhAL/SctVL, with a groove on the adjacent protomer's cytoplasmic domain, which stabilises the nonameric ring assembly. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11820.map.gz emd_11820.map.gz | 478.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11820-v30.xml emd-11820-v30.xml emd-11820.xml emd-11820.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11820_fsc.xml emd_11820_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_11820.png emd_11820.png | 165.4 KB | ||
| Masks |  emd_11820_msk_1.map emd_11820_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11820.cif.gz emd-11820.cif.gz | 6.4 KB | ||
| Others |  emd_11820_half_map_1.map.gz emd_11820_half_map_1.map.gz emd_11820_half_map_2.map.gz emd_11820_half_map_2.map.gz | 410.6 MB 410.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11820 http://ftp.pdbj.org/pub/emdb/structures/EMD-11820 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11820 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11820 | HTTPS FTP |
-Related structure data
| Related structure data |  7alwMC  7amyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11820.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11820.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the SctV cytoplasmic domain | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11820_msk_1.map emd_11820_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_11820_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_11820_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Double nonamer of the SctV cytoplasmic domain from Yersinia enter...
| Entire | Name: Double nonamer of the SctV cytoplasmic domain from Yersinia enterocolitica |
|---|---|
| Components |
|
-Supramolecule #1: Double nonamer of the SctV cytoplasmic domain from Yersinia enter...
| Supramolecule | Name: Double nonamer of the SctV cytoplasmic domain from Yersinia enterocolitica type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) |
| Molecular weight | Theoretical: 1.4 MDa |
-Macromolecule #1: Low calcium response protein
| Macromolecule | Name: Low calcium response protein / type: protein_or_peptide / ID: 1 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) |
| Molecular weight | Theoretical: 78.667281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNPHDLEWLN RIGERKDIML AVLLLAVVFM MVLPLPPIVL DILIAVNMTI SVVLLMIAIY INSPLQFSAF PAVLLVTTLF RLALSVSTT RMILLQADAG QIVYTFGNFV VGGNLIVGIV IFLIITIVQF LVITKGSERV AEVSARFSLD AMPGKQMSID G DMRAGVID ...String: MNPHDLEWLN RIGERKDIML AVLLLAVVFM MVLPLPPIVL DILIAVNMTI SVVLLMIAIY INSPLQFSAF PAVLLVTTLF RLALSVSTT RMILLQADAG QIVYTFGNFV VGGNLIVGIV IFLIITIVQF LVITKGSERV AEVSARFSLD AMPGKQMSID G DMRAGVID VNEARERRAT IEKESQMFGS MDGAMKFVKG DAIAGLIIIF VNILGGVTIG VTQKGLAAAE ALQLYSILTV GD GMVSQVP ALLIAITAGI IVTRVSSEDS SDLGSDIGKQ VVAQPKAMLI GGVLLLLFGL IPGFPTVTFL ILALLVGCGG YML SRKQSR NDEANQDLQS ILTSGSGAPA ARTKAKTSGA NKGRLGEQEA FAMTVPLLID VDSSQQEALE AIALNDELVR VRRA LYLDL GVPFPGIHLR FNEGMGEGEY LISLQEVPVA RGELKAGYLL VRESVSQLEL LGIPYEKGEH LLPDQETFWV SVEYE ERLE KSQLEFFSHS QVLTWHLSHV LREYAEDFIG IQETRYLLEQ MEGGYGELIK EVQRIVPLQR MTEILQRLVG EDISIR NMR SILEAMVEWG QKEKDVVQLT EYIRSSLKRY ICYKYANGNN ILPAYLFDQE VEEKIRSGVR QTSAGSYLAL DPAVTES LL EQVRKTIGDL SQIQSKPVLI VSMDIRRYVR KLIESEYYGL PVLSYQELTQ QINIQPLGRV CLENLYFQ UniProtKB: Low calcium response protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)