[English] 日本語
 Yorodumi
Yorodumi- EMDB-11780: Apo-state type 3 secretion system export apparatus complex from S... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11780 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Apo-state type 3 secretion system export apparatus complex from Salmonella enterica typhimurium | |||||||||
 Map data Map data | Postprocess Bfactor: -30 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | T3SS / Export Apparatus / Injectisome / Needle Complex / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationThe IPAF inflammasome / type III protein secretion system complex / protein secretion by the type III secretion system / bacterial-type flagellum-dependent swarming motility / bacterial-type flagellum assembly / protein secretion / protein targeting / protein transport / cell surface / extracellular region ...The IPAF inflammasome / type III protein secretion system complex / protein secretion by the type III secretion system / bacterial-type flagellum-dependent swarming motility / bacterial-type flagellum assembly / protein secretion / protein targeting / protein transport / cell surface / extracellular region / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) / Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) /  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) | |||||||||
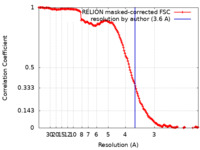
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Marlovits T | |||||||||
| Funding support |  Austria, Austria,  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation. Authors: Sean Miletic / Dirk Fahrenkamp / Nikolaus Goessweiner-Mohr / Jiri Wald / Maurice Pantel / Oliver Vesper / Vadim Kotov / Thomas C Marlovits /   Abstract: Many bacterial pathogens rely on virulent type III secretion systems (T3SSs) or injectisomes to translocate effector proteins in order to establish infection. The central component of the injectisome ...Many bacterial pathogens rely on virulent type III secretion systems (T3SSs) or injectisomes to translocate effector proteins in order to establish infection. The central component of the injectisome is the needle complex which assembles a continuous conduit crossing the bacterial envelope and the host cell membrane to mediate effector protein translocation. However, the molecular principles underlying type III secretion remain elusive. Here, we report a structure of an active Salmonella enterica serovar Typhimurium needle complex engaged with the effector protein SptP in two functional states, revealing the complete 800Å-long secretion conduit and unraveling the critical role of the export apparatus (EA) subcomplex in type III secretion. Unfolded substrates enter the EA through a hydrophilic constriction formed by SpaQ proteins, which enables side chain-independent substrate transport. Above, a methionine gasket formed by SpaP proteins functions as a gate that dilates to accommodate substrates while preventing leaky pore formation. Following gate penetration, a moveable SpaR loop first folds up to then support substrate transport. Together, these findings establish the molecular basis for substrate translocation through T3SSs and improve our understanding of bacterial pathogenicity and motility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11780.map.gz emd_11780.map.gz | 283.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11780-v30.xml emd-11780-v30.xml emd-11780.xml emd-11780.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11780_fsc.xml emd_11780_fsc.xml | 15.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_11780.png emd_11780.png | 50.3 KB | ||
| Filedesc metadata |  emd-11780.cif.gz emd-11780.cif.gz | 5.6 KB | ||
| Others |  emd_11780_additional_1.map.gz emd_11780_additional_1.map.gz emd_11780_half_map_1.map.gz emd_11780_half_map_1.map.gz emd_11780_half_map_2.map.gz emd_11780_half_map_2.map.gz | 244.1 MB 245.5 MB 245.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11780 http://ftp.pdbj.org/pub/emdb/structures/EMD-11780 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11780 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11780 | HTTPS FTP |
-Related structure data
| Related structure data |  7agxMC  7ah9C  7ahiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11780.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11780.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocess Bfactor: -30 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
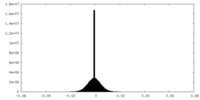
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Refine3D
| File | emd_11780_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refine3D | ||||||||||||
| Projections & Slices |
| ||||||||||||
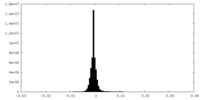
| Density Histograms |
-Half map: #1
| File | emd_11780_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
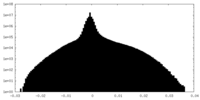
| Density Histograms |
-Half map: #2
| File | emd_11780_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
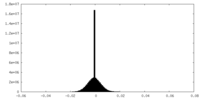
| Density Histograms |
- Sample components
Sample components
-Entire : Export apparatus core complex with inner rod and needle filament ...
| Entire | Name: Export apparatus core complex with inner rod and needle filament proteins PrgJ and PrgI. |
|---|---|
| Components |
|
-Supramolecule #1: Export apparatus core complex with inner rod and needle filament ...
| Supramolecule | Name: Export apparatus core complex with inner rod and needle filament proteins PrgJ and PrgI. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Apo-state |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria)Location in cell: Membrane |
| Molecular weight | Theoretical: 336 KDa |
-Macromolecule #1: Surface presentation of antigens protein SpaP
| Macromolecule | Name: Surface presentation of antigens protein SpaP / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) |
| Molecular weight | Theoretical: 25.249596 KDa |
| Sequence | String: MGNDISLIAL LAFSTLLPFI IASGTCFVKF SIVFVMVRNA LGLQQIPSNM TLNGVALLLS MFVMWPIMHD AYVYFEDEDV TFNDISSLS KHVDEGLDGY RDYLIKYSDR ELVQFFENAQ LKRQYGEETE TVKRDKDEIE KPSIFALLPA YALSEIKSAF K IGFYLYLP ...String: MGNDISLIAL LAFSTLLPFI IASGTCFVKF SIVFVMVRNA LGLQQIPSNM TLNGVALLLS MFVMWPIMHD AYVYFEDEDV TFNDISSLS KHVDEGLDGY RDYLIKYSDR ELVQFFENAQ LKRQYGEETE TVKRDKDEIE KPSIFALLPA YALSEIKSAF K IGFYLYLP FVVVDLVVSS VLLALGMMMM SPVTISTPIK LVLFVALDGW TLLSKGLILQ YMDIAT UniProtKB: Surface presentation of antigens protein SpaP |
-Macromolecule #2: Surface presentation of antigens protein SpaR
| Macromolecule | Name: Surface presentation of antigens protein SpaR / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) |
| Molecular weight | Theoretical: 28.499533 KDa |
| Sequence | String: MFYALYFEIH HLVASAALGF ARVAPIFFFL PFLNSGVLSG APRNAIIILV ALGVWPHALN EAPPFLSVAM IPLVLQEAAV GVMLGCLLS WPFWVMHALG CIIDNQRGAT LSSSIDPANG IDTSEMANFL NMFAAVVYLQ NGGLVTMVDV LNKSYQLCDP M NECTPSLP ...String: MFYALYFEIH HLVASAALGF ARVAPIFFFL PFLNSGVLSG APRNAIIILV ALGVWPHALN EAPPFLSVAM IPLVLQEAAV GVMLGCLLS WPFWVMHALG CIIDNQRGAT LSSSIDPANG IDTSEMANFL NMFAAVVYLQ NGGLVTMVDV LNKSYQLCDP M NECTPSLP PLLTFINQVA QNALVLASPV VLVLLLSEVF LGLLSRFAPQ MNAFAISLTV KSGIAVLIML LYFSPVLPDN VL RLSFQAT GLSSWFYERG ATHVLE UniProtKB: Surface presentation of antigens protein SpaR |
-Macromolecule #3: Surface presentation of antigens protein SpaQ
| Macromolecule | Name: Surface presentation of antigens protein SpaQ / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) |
| Molecular weight | Theoretical: 9.363229 KDa |
| Sequence | String: MDDLVFAGNK ALYLVLILSG WPTIVATIIG LLVGLFQTVT QLQEQTLPFG IKLLGVCLCL FLLSGWYGEV LLSYGRQVIF LALAKG UniProtKB: Surface presentation of antigens protein SpaQ |
-Macromolecule #4: Protein PrgJ
| Macromolecule | Name: Protein PrgJ / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) |
| Molecular weight | Theoretical: 10.934425 KDa |
| Sequence | String: MSIATIVPEN AVIGQAVNIR SMETDIVSLD DRLLQAFSGS AIATAVDKQT ITNRIEDPNL VTDPKELAIS QEMISDYNLY VSMVSTLTR KGVGAVETLL RS UniProtKB: Protein PrgJ |
-Macromolecule #5: Protein PrgI
| Macromolecule | Name: Protein PrgI / type: protein_or_peptide / ID: 5 / Number of copies: 17 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) |
| Molecular weight | Theoretical: 8.864868 KDa |
| Sequence | String: MATPWSGYLD DVSAKFDTGV DNLQTQVTEA LDKLAAKPSD PALLAAYQSK LSEYNLYRNA QSNTVKVFKD IDAAIIQNFR UniProtKB: SPI-1 type 3 secretion system needle filament protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 31.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7agx: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)