+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11561 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | mechanosensitive channel YnaI in an intermediate conformation | |||||||||
 Map data Map data | map from Relion Post-process, sharpened with a B-factor of -134, without map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
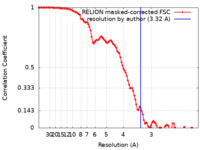
| Method | single particle reconstruction / cryo EM / Resolution: 3.32 Å | |||||||||
 Authors Authors | Flegler VJ / Rasmussen A / Rao S / Wu N / Zenobi R / Sansom MSP / Hedrich R / Rasmussen T / Boettcher B | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: The MscS-like channel YnaI has a gating mechanism based on flexible pore helices. Authors: Vanessa Judith Flegler / Akiko Rasmussen / Shanlin Rao / Na Wu / Renato Zenobi / Mark S P Sansom / Rainer Hedrich / Tim Rasmussen / Bettina Böttcher /    Abstract: The mechanosensitive channel of small conductance (MscS) is the prototype of an evolutionarily diversified large family that fine-tunes osmoregulation but is likely to fulfill additional functions. ...The mechanosensitive channel of small conductance (MscS) is the prototype of an evolutionarily diversified large family that fine-tunes osmoregulation but is likely to fulfill additional functions. has six osmoprotective paralogs with different numbers of transmembrane helices. These helices are important for gating and sensing in MscS but the role of the additional helices in the paralogs is not understood. The medium-sized channel YnaI was extracted and delivered in native nanodiscs in closed-like and open-like conformations using the copolymer diisobutylene/maleic acid (DIBMA) for structural studies. Here we show by electron cryomicroscopy that YnaI has an extended sensor paddle that during gating relocates relative to the pore concomitant with bending of a GGxGG motif in the pore helices. YnaI is the only one of the six paralogs that has this GGxGG motif allowing the sensor paddle to move outward. Access to the pore is through a vestibule on the cytosolic side that is fenestrated by side portals. In YnaI, these portals are obstructed by aromatic side chains but are still fully hydrated and thus support conductance. For comparison with large-sized channels, we determined the structure of YbiO, which showed larger portals and a wider pore with no GGxGG motif. Further in silico comparison of MscS, YnaI, and YbiO highlighted differences in the hydrophobicity and wettability of their pores and vestibule interiors. Thus, MscS-like channels of different sizes have a common core architecture but show different gating mechanisms and fine-tuned conductive properties. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11561.map.gz emd_11561.map.gz | 28.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11561-v30.xml emd-11561-v30.xml emd-11561.xml emd-11561.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11561_fsc.xml emd_11561_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11561.png emd_11561.png | 124.9 KB | ||
| Masks |  emd_11561_msk_1.map emd_11561_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Others |  emd_11561_additional_1.map.gz emd_11561_additional_1.map.gz emd_11561_half_map_1.map.gz emd_11561_half_map_1.map.gz emd_11561_half_map_2.map.gz emd_11561_half_map_2.map.gz | 28 MB 28.2 MB 28.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11561 http://ftp.pdbj.org/pub/emdb/structures/EMD-11561 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11561 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11561 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11561.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11561.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map from Relion Post-process, sharpened with a B-factor of -134, without map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0635 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
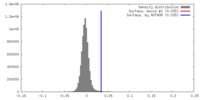
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11561_msk_1.map emd_11561_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: raw map: from Relion Refine3D
| File | emd_11561_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | raw map: from Relion Refine3D | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_11561_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_11561_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : YnaI in an intermediate conformation
| Entire | Name: YnaI in an intermediate conformation |
|---|---|
| Components |
|
-Supramolecule #1: YnaI in an intermediate conformation
| Supramolecule | Name: YnaI in an intermediate conformation / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: YnaI was purified with the detergent DDM and reconstituted into liposomes. After addition of LPC it was extracted using DIBMA co-polymer. |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 278 KDa |
-Macromolecule #1: YnaI
| Macromolecule | Name: YnaI / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MIAELFTNNA LNLVIIFGSC AALILMSFWF RRGNRKRKGF LFHAVQFLIY TIIISAVGSI INYVIENYKL KFITPGVID FICTSLIAVI LTIKLFLLIN QFEKQQIKKG RDITSARIMS RIIKITIIVV LVLLYGEHFG M SLSGLLTF AGIAGLAVGM AGKDILSNFF ...String: MIAELFTNNA LNLVIIFGSC AALILMSFWF RRGNRKRKGF LFHAVQFLIY TIIISAVGSI INYVIENYKL KFITPGVID FICTSLIAVI LTIKLFLLIN QFEKQQIKKG RDITSARIMS RIIKITIIVV LVLLYGEHFG M SLSGLLTF AGIAGLAVGM AGKDILSNFF SGIMLYFDRP FSIGDWIRSP DRNIEGTVAE IGWRITKIKT FD NRPLYVP NSLFSSISVE NPGRMTNRRI TTTIGLRYED AAKVGVIVEA VREMLKNHPA IDQRQTLLVY FNQ FADSSL NIMVYCFTKT TVWAEWLAAQ QDVYLKIIDI VQSHGADFAF PSQTLYMDNI TPPDQGRLEH HHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 4161 / Average exposure time: 63.0 sec. / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)