+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11385 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Treponema denticola chemotaxis signalling arrays | |||||||||
 Map data Map data | Treponema denticola chemotaxis arrays | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Treponema denticola (bacteria) Treponema denticola (bacteria) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Muok AR / Yang W / Briegel A | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Atypical chemoreceptor arrays accommodate high membrane curvature. Authors: Alise R Muok / Davi R Ortega / Kurni Kurniyati / Wen Yang / Zachary A Maschmann / Adam Sidi Mabrouk / Chunhao Li / Brian R Crane / Ariane Briegel /   Abstract: The prokaryotic chemotaxis system is arguably the best-understood signaling pathway in biology. In all previously described species, chemoreceptors organize into a hexagonal (P6 symmetry) extended ...The prokaryotic chemotaxis system is arguably the best-understood signaling pathway in biology. In all previously described species, chemoreceptors organize into a hexagonal (P6 symmetry) extended array. Here, we report an alternative symmetry (P2) of the chemotaxis apparatus that emerges from a strict linear organization of the histidine kinase CheA in Treponema denticola cells, which possesses arrays with the highest native curvature investigated thus far. Using cryo-ET, we reveal that Td chemoreceptor arrays assume an unusual arrangement of the supra-molecular protein assembly that has likely evolved to accommodate the high membrane curvature. The arrays have several atypical features, such as an extended dimerization domain of CheA and a variant CheW-CheR-like fusion protein that is critical for maintaining an ordered chemosensory apparatus. Furthermore, the previously characterized Td oxygen sensor ODP influences CheA ordering. These results suggest a greater diversity of the chemotaxis signaling system than previously thought. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11385.map.gz emd_11385.map.gz | 2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11385-v30.xml emd-11385-v30.xml emd-11385.xml emd-11385.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11385_fsc.xml emd_11385_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11385.png emd_11385.png | 31 KB | ||
| Masks |  emd_11385_msk_1.map emd_11385_msk_1.map | 8 MB |  Mask map Mask map | |
| Others |  emd_11385_half_map_1.map.gz emd_11385_half_map_1.map.gz emd_11385_half_map_2.map.gz emd_11385_half_map_2.map.gz | 7.4 MB 7.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11385 http://ftp.pdbj.org/pub/emdb/structures/EMD-11385 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11385 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11385 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11385.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11385.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Treponema denticola chemotaxis arrays | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.026 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11385_msk_1.map emd_11385_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
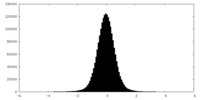
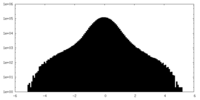
| Density Histograms |
-Half map: Half map 2
| File | emd_11385_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_11385_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Chemotaxis arrays in a Treponema denticola ODP (TDE2498) deletion...
| Entire | Name: Chemotaxis arrays in a Treponema denticola ODP (TDE2498) deletion mutant |
|---|---|
| Components |
|
-Supramolecule #1: Chemotaxis arrays in a Treponema denticola ODP (TDE2498) deletion...
| Supramolecule | Name: Chemotaxis arrays in a Treponema denticola ODP (TDE2498) deletion mutant type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Treponema denticola (bacteria) Treponema denticola (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 20 K / Instrument: LEICA PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




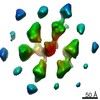







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)