+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11257 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Native complex I, open1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / respiration / NADH / proton pump / mitochondria / iron-sulphur cluster / oxidoreductase / membrane protein / ELECTRON TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology information: / : / ubiquinone binding / acyl binding / electron transport coupled proton transport / NADH:ubiquinone reductase (H+-translocating) / acyl carrier activity / apoptotic mitochondrial changes / NADH dehydrogenase activity / mitochondrial respiratory chain complex I assembly ...: / : / ubiquinone binding / acyl binding / electron transport coupled proton transport / NADH:ubiquinone reductase (H+-translocating) / acyl carrier activity / apoptotic mitochondrial changes / NADH dehydrogenase activity / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / respiratory chain complex I / membrane => GO:0016020 / NADH dehydrogenase (ubiquinone) activity / ATP metabolic process / ATP synthesis coupled electron transport / reactive oxygen species metabolic process / regulation of mitochondrial membrane potential / respiratory electron transport chain / circadian rhythm / electron transport chain / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / NAD binding / FMN binding / 4 iron, 4 sulfur cluster binding / response to oxidative stress / mitochondrial inner membrane / mitochondrial matrix / protein-containing complex binding / mitochondrion / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
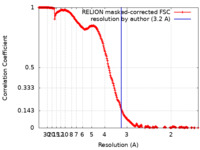
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Kampjut D / Sazanov LA | |||||||||
| Funding support | European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: The coupling mechanism of mammalian respiratory complex I. Authors: Domen Kampjut / Leonid A Sazanov /  Abstract: Mitochondrial complex I couples NADH:ubiquinone oxidoreduction to proton pumping by an unknown mechanism. Here, we present cryo-electron microscopy structures of ovine complex I in five different ...Mitochondrial complex I couples NADH:ubiquinone oxidoreduction to proton pumping by an unknown mechanism. Here, we present cryo-electron microscopy structures of ovine complex I in five different conditions, including turnover, at resolutions up to 2.3 to 2.5 angstroms. Resolved water molecules allowed us to experimentally define the proton translocation pathways. Quinone binds at three positions along the quinone cavity, as does the inhibitor rotenone that also binds within subunit ND4. Dramatic conformational changes around the quinone cavity couple the redox reaction to proton translocation during open-to-closed state transitions of the enzyme. In the induced deactive state, the open conformation is arrested by the ND6 subunit. We propose a detailed molecular coupling mechanism of complex I, which is an unexpected combination of conformational changes and electrostatic interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11257.map.gz emd_11257.map.gz | 58.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11257-v30.xml emd-11257-v30.xml emd-11257.xml emd-11257.xml | 64.3 KB 64.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11257_fsc.xml emd_11257_fsc.xml | 22.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_11257.png emd_11257.png | 82.1 KB | ||
| Filedesc metadata |  emd-11257.cif.gz emd-11257.cif.gz | 14.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11257 http://ftp.pdbj.org/pub/emdb/structures/EMD-11257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11257 | HTTPS FTP |
-Related structure data
| Related structure data |  6zkpMC  6zk9C  6zkaC  6zkbC  6zkcC  6zkdC  6zkeC  6zkfC  6zkgC  6zkhC  6zkiC  6zkjC  6zkkC  6zklC  6zkmC  6zknC  6zkoC  6zkqC  6zkrC  6zksC  6zktC  6zkuC  6zkvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11257.map.gz / Format: CCP4 / Size: 63.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11257.map.gz / Format: CCP4 / Size: 63.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
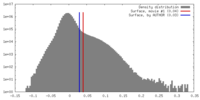
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Native complex I, open1
+Supramolecule #1: Native complex I, open1
+Macromolecule #1: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial
+Macromolecule #2: Mitochondrial complex I, 24 kDa subunit
+Macromolecule #3: NADH:ubiquinone oxidoreductase core subunit S1
+Macromolecule #4: Mitochondrial complex I, 49 kDa subunit
+Macromolecule #5: NADH:ubiquinone oxidoreductase core subunit S3
+Macromolecule #6: Mitochondrial complex I, PSST subunit
+Macromolecule #7: Mitochondrial complex I, TYKY subunit
+Macromolecule #8: NADH-ubiquinone oxidoreductase chain 3
+Macromolecule #9: NADH-ubiquinone oxidoreductase chain 1
+Macromolecule #10: NADH-ubiquinone oxidoreductase chain 6
+Macromolecule #11: NADH-ubiquinone oxidoreductase chain 4L
+Macromolecule #12: NADH-ubiquinone oxidoreductase chain 5
+Macromolecule #13: NADH-ubiquinone oxidoreductase chain 4
+Macromolecule #14: NADH-ubiquinone oxidoreductase chain 2
+Macromolecule #15: Mitochondrial complex I, B14.7 subunit
+Macromolecule #16: NADH:ubiquinone oxidoreductase subunit B5
+Macromolecule #17: Acyl carrier protein
+Macromolecule #18: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8
+Macromolecule #19: Mitochondrial complex I, PDSW subunit
+Macromolecule #20: Mitochondrial complex I, 10 kDa subunit
+Macromolecule #21: Mitochondrial complex I, 13 kDa subunit
+Macromolecule #22: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial
+Macromolecule #23: NADH:ubiquinone oxidoreductase subunit A9
+Macromolecule #24: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2
+Macromolecule #25: Mitochondrial complex I, B13 subunit
+Macromolecule #26: NADH:ubiquinone oxidoreductase subunit A6
+Macromolecule #27: Mitochondrial complex I, B14.5a subunit
+Macromolecule #28: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12
+Macromolecule #29: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mi...
+Macromolecule #30: NADH:ubiquinone oxidoreductase subunit S5
+Macromolecule #31: NADH:ubiquinone oxidoreductase subunit A3
+Macromolecule #32: NADH:ubiquinone oxidoreductase subunit B3
+Macromolecule #33: NADH dehydrogenase [ubiquinone] 1 subunit C2
+Macromolecule #34: NADH:ubiquinone oxidoreductase subunit B4
+Macromolecule #35: Mitochondrial complex I, B16.6 subunit
+Macromolecule #36: Mitochondrial complex I, B17 subunit
+Macromolecule #37: NADH:ubiquinone oxidoreductase subunit B7
+Macromolecule #38: NADH:ubiquinone oxidoreductase subunit B9
+Macromolecule #39: NADH:ubiquinone oxidoreductase subunit B2
+Macromolecule #40: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mito...
+Macromolecule #41: Mitochondrial complex I, ESSS subunit
+Macromolecule #42: Mitochondrial complex I, KFYI subunit
+Macromolecule #43: Mitochondrial complex I, MNLL subunit
+Macromolecule #44: Mitochondrial complex I, MWFE subunit
+Macromolecule #45: IRON/SULFUR CLUSTER
+Macromolecule #46: FLAVIN MONONUCLEOTIDE
+Macromolecule #47: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #48: POTASSIUM ION
+Macromolecule #49: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #50: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #51: CARDIOLIPIN
+Macromolecule #52: S-[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-b...
+Macromolecule #53: ZINC ION
+Macromolecule #54: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
+Macromolecule #55: ADENOSINE MONOPHOSPHATE
+Macromolecule #56: MYRISTIC ACID
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 100.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)