[English] 日本語
 Yorodumi
Yorodumi- EMDB-11235: Respiratory complex I from Thermus thermophilus, NAD+ dataset, ma... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11235 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Respiratory complex I from Thermus thermophilus, NAD+ dataset, major state | |||||||||
 Map data Map data | Postprocessed map, NAD dataset, major state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Respiratory chain / complex I / NADH:ubiquinone oxidoreductase / electron transfer / proton translocation / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTranslocases; Catalysing the translocation of protons; Linked to oxidoreductase reactions / NADH dehydrogenase (quinone) (non-electrogenic) activity / NADH dehydrogenase complex / molybdopterin cofactor binding / oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor / NADH dehydrogenase activity / iron-sulfur cluster assembly / ubiquinone binding / electron transport coupled proton transport / respiratory chain complex I ...Translocases; Catalysing the translocation of protons; Linked to oxidoreductase reactions / NADH dehydrogenase (quinone) (non-electrogenic) activity / NADH dehydrogenase complex / molybdopterin cofactor binding / oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor / NADH dehydrogenase activity / iron-sulfur cluster assembly / ubiquinone binding / electron transport coupled proton transport / respiratory chain complex I / NADH dehydrogenase (ubiquinone) activity / quinone binding / ATP synthesis coupled electron transport / ferric iron binding / aerobic respiration / 2 iron, 2 sulfur cluster binding / NAD binding / FMN binding / 4 iron, 4 sulfur cluster binding / iron ion binding / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | |||||||||
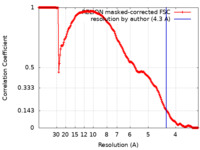
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Kaszuba K / Tambalo M | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Key role of quinone in the mechanism of respiratory complex I. Authors: Javier Gutiérrez-Fernández / Karol Kaszuba / Gurdeep S Minhas / Rozbeh Baradaran / Margherita Tambalo / David T Gallagher / Leonid A Sazanov /    Abstract: Complex I is the first and the largest enzyme of respiratory chains in bacteria and mitochondria. The mechanism which couples spatially separated transfer of electrons to proton translocation in ...Complex I is the first and the largest enzyme of respiratory chains in bacteria and mitochondria. The mechanism which couples spatially separated transfer of electrons to proton translocation in complex I is not known. Here we report five crystal structures of T. thermophilus enzyme in complex with NADH or quinone-like compounds. We also determined cryo-EM structures of major and minor native states of the complex, differing in the position of the peripheral arm. Crystal structures show that binding of quinone-like compounds (but not of NADH) leads to a related global conformational change, accompanied by local re-arrangements propagating from the quinone site to the nearest proton channel. Normal mode and molecular dynamics analyses indicate that these are likely to represent the first steps in the proton translocation mechanism. Our results suggest that quinone binding and chemistry play a key role in the coupling mechanism of complex I. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11235.map.gz emd_11235.map.gz | 479.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11235-v30.xml emd-11235-v30.xml emd-11235.xml emd-11235.xml | 30.6 KB 30.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11235_fsc.xml emd_11235_fsc.xml | 18 KB | Display |  FSC data file FSC data file |
| Images |  emd_11235.png emd_11235.png | 212.9 KB | ||
| Filedesc metadata |  emd-11235.cif.gz emd-11235.cif.gz | 9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11235 http://ftp.pdbj.org/pub/emdb/structures/EMD-11235 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11235 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11235 | HTTPS FTP |
-Validation report
| Summary document |  emd_11235_validation.pdf.gz emd_11235_validation.pdf.gz | 712.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11235_full_validation.pdf.gz emd_11235_full_validation.pdf.gz | 712.3 KB | Display | |
| Data in XML |  emd_11235_validation.xml.gz emd_11235_validation.xml.gz | 15.7 KB | Display | |
| Data in CIF |  emd_11235_validation.cif.gz emd_11235_validation.cif.gz | 22.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11235 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11235 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11235 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11235 | HTTPS FTP |
-Related structure data
| Related structure data |  6zjlMC  6i0dC  6i1pC  6q8oC  6q8wC  6q8xC  6y11C  6ziyC  6zjnC  6zjyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11235.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11235.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map, NAD dataset, major state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.72 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Respiratory complex I from Thermus thermophilus
+Supramolecule #1: Respiratory complex I from Thermus thermophilus
+Macromolecule #1: NADH-quinone oxidoreductase subunit 1
+Macromolecule #2: NADH-quinone oxidoreductase subunit 2
+Macromolecule #3: NADH-quinone oxidoreductase subunit 3
+Macromolecule #4: NADH-quinone oxidoreductase subunit 4
+Macromolecule #5: NADH-quinone oxidoreductase subunit 5
+Macromolecule #6: NADH-quinone oxidoreductase subunit 6
+Macromolecule #7: NADH-quinone oxidoreductase subunit 9
+Macromolecule #8: NADH-quinone oxidoreductase subunit 15
+Macromolecule #9: NADH-quinone oxidoreductase subunit 7
+Macromolecule #10: NADH-quinone oxidoreductase subunit 10
+Macromolecule #11: NADH-quinone oxidoreductase subunit 11
+Macromolecule #12: NADH-quinone oxidoreductase subunit 12
+Macromolecule #13: NADH-quinone oxidoreductase subunit 13
+Macromolecule #14: NADH-quinone oxidoreductase subunit 14
+Macromolecule #15: NADH-quinone oxidoreductase subunit 8
+Macromolecule #16: IRON/SULFUR CLUSTER
+Macromolecule #17: FLAVIN MONONUCLEOTIDE
+Macromolecule #18: FE2/S2 (INORGANIC) CLUSTER
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.0 mg/mL |
|---|---|
| Buffer | pH: 6 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK III |
| Details | with 5 mM NAD+ |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-34 / Number grids imaged: 1 / Number real images: 710 / Average exposure time: 2.0 sec. / Average electron dose: 34.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)