[English] 日本語
 Yorodumi
Yorodumi- EMDB-11131: Subtomogram average of elastase treated human Uromodulin filament... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11131 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
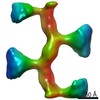
| Title | Subtomogram average of elastase treated human Uromodulin filaments (eUmod) | ||||||||||||||||||||||||
 Map data Map data | Subtomogram average of elastase treated human Uromodulin (eUmod) | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 17.3 Å | ||||||||||||||||||||||||
 Authors Authors | Weiss GL / Stanisich JJ / Sauer MM / Lin C-W / Eras J / Zyla DS / Trueck J / Devuyst O / Aebi M / Pilhofer M / Glockshuber R | ||||||||||||||||||||||||
| Funding support |  Switzerland, 7 items Switzerland, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Architecture and function of human uromodulin filaments in urinary tract infections. Authors: Gregor L Weiss / Jessica J Stanisich / Maximilian M Sauer / Chia-Wei Lin / Jonathan Eras / Dawid S Zyla / Johannes Trück / Olivier Devuyst / Markus Aebi / Martin Pilhofer / Rudi Glockshuber /   Abstract: Uromodulin is the most abundant protein in human urine, and it forms filaments that antagonize the adhesion of uropathogens; however, the filament structure and mechanism of protection remain poorly ...Uromodulin is the most abundant protein in human urine, and it forms filaments that antagonize the adhesion of uropathogens; however, the filament structure and mechanism of protection remain poorly understood. We used cryo-electron tomography to show that the uromodulin filament consists of a zigzag-shaped backbone with laterally protruding arms. N-glycosylation mapping and biophysical assays revealed that uromodulin acts as a multivalent ligand for the bacterial type 1 pilus adhesin, presenting specific epitopes on the regularly spaced arms. Imaging of uromodulin-uropathogen interactions in vitro and in patient urine showed that uromodulin filaments associate with uropathogens and mediate bacterial aggregation, which likely prevents adhesion and allows clearance by micturition. These results provide a framework for understanding uromodulin in urinary tract infections and in its more enigmatic roles in physiology and disease. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11131.map.gz emd_11131.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11131-v30.xml emd-11131-v30.xml emd-11131.xml emd-11131.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11131.png emd_11131.png | 23.2 KB | ||
| Masks |  emd_11131_msk_1.map emd_11131_msk_1.map | 501 KB |  Mask map Mask map | |
| Others |  emd_11131_half_map_1.map.gz emd_11131_half_map_1.map.gz emd_11131_half_map_2.map.gz emd_11131_half_map_2.map.gz | 270.8 KB 269 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11131 http://ftp.pdbj.org/pub/emdb/structures/EMD-11131 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11131 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11131 | HTTPS FTP |
-Validation report
| Summary document |  emd_11131_validation.pdf.gz emd_11131_validation.pdf.gz | 378.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11131_full_validation.pdf.gz emd_11131_full_validation.pdf.gz | 377.9 KB | Display | |
| Data in XML |  emd_11131_validation.xml.gz emd_11131_validation.xml.gz | 7.4 KB | Display | |
| Data in CIF |  emd_11131_validation.cif.gz emd_11131_validation.cif.gz | 8.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11131 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11131 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11131 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11131 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11131.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11131.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of elastase treated human Uromodulin (eUmod) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.449 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11131_msk_1.map emd_11131_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map for FSC calculation (even)
| File | emd_11131_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map for FSC calculation (even) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map for FSC calculation (odd)
| File | emd_11131_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map for FSC calculation (odd) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : elastase treated human Uromodulin filaments
| Entire | Name: elastase treated human Uromodulin filaments |
|---|---|
| Components |
|
-Supramolecule #1: elastase treated human Uromodulin filaments
| Supramolecule | Name: elastase treated human Uromodulin filaments / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 17.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number subtomograms used: 2177 |
|---|---|
| Extraction | Number tomograms: 7 / Number images used: 6646 |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)