+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11089 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Transcription termination intermediate complex 3 | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-templated transcription elongation / ATP-dependent activity, acting on RNA / transcription elongation-coupled chromatin remodeling / RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / protein complex oligomerization / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding ...DNA-templated transcription elongation / ATP-dependent activity, acting on RNA / transcription elongation-coupled chromatin remodeling / RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / protein complex oligomerization / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / nitrate assimilation / translation elongation factor activity / DNA-directed RNA polymerase complex / regulation of DNA-templated transcription elongation / transcription elongation factor complex / transcription antitermination / cell motility / DNA-templated transcription initiation / DNA-templated transcription termination / helicase activity / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / ribosome biogenesis / response to heat / protein-containing complex assembly / intracellular iron ion homeostasis / protein dimerization activity / DNA-binding transcription factor activity / protein domain specific binding / response to antibiotic / nucleotide binding / DNA-templated transcription / magnesium ion binding / ATP hydrolysis activity / DNA binding / RNA binding / zinc ion binding / ATP binding / identical protein binding / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
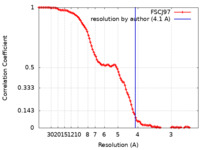
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||||||||
 Authors Authors | Said N / Hilal T / Buerger J / Mielke T / Loll B / Wahl MC | |||||||||||||||
| Funding support |  Germany, Germany,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase ρ. Authors: Nelly Said / Tarek Hilal / Nicholas D Sunday / Ajay Khatri / Jörg Bürger / Thorsten Mielke / Georgiy A Belogurov / Bernhard Loll / Ranjan Sen / Irina Artsimovitch / Markus C Wahl /     Abstract: Factor-dependent transcription termination mechanisms are poorly understood. We determined a series of cryo-electron microscopy structures portraying the hexameric adenosine triphosphatase (ATPase) ...Factor-dependent transcription termination mechanisms are poorly understood. We determined a series of cryo-electron microscopy structures portraying the hexameric adenosine triphosphatase (ATPase) ρ on a pathway to terminating NusA/NusG-modified elongation complexes. An open ρ ring contacts NusA, NusG, and multiple regions of RNA polymerase, trapping and locally unwinding proximal upstream DNA. NusA wedges into the ρ ring, initially sequestering RNA. Upon deflection of distal upstream DNA over the RNA polymerase zinc-binding domain, NusA rotates underneath one capping ρ subunit, which subsequently captures RNA. After detachment of NusG and clamp opening, RNA polymerase loses its grip on the RNA:DNA hybrid and is inactivated. Our structural and functional analyses suggest that ρ, and other termination factors across life, may use analogous strategies to allosterically trap transcription complexes in a moribund state. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11089.map.gz emd_11089.map.gz | 3.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11089-v30.xml emd-11089-v30.xml emd-11089.xml emd-11089.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11089_fsc.xml emd_11089_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_11089.png emd_11089.png | 94.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11089 http://ftp.pdbj.org/pub/emdb/structures/EMD-11089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11089 | HTTPS FTP |
-Related structure data
| Related structure data |  6z9rMC  6z9pC  6z9qC  6z9sC  6z9tC  7adbC  7adcC  7addC  7adeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11089.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11089.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Transcription termination complex
| Entire | Name: Transcription termination complex |
|---|---|
| Components |
|
-Supramolecule #1: Transcription termination complex
| Supramolecule | Name: Transcription termination complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 800 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 10.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 25.0 µm / Nominal defocus min: 8.0 µm / Nominal magnification: 31000 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)