[English] 日本語
 Yorodumi
Yorodumi- EMDB-11067: CryoEM structure of horse sodium/proton exchanger NHE9 without C-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11067 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of horse sodium/proton exchanger NHE9 without C-terminal regulatory domain in an inward-facing conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRANSPORT PROTEIN / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphagosome maturation / potassium:proton antiporter activity / sodium:proton antiporter activity / early phagosome / sodium ion import across plasma membrane / potassium ion transmembrane transport / proton transmembrane transport / sodium ion transmembrane transport / regulation of intracellular pH / recycling endosome ...phagosome maturation / potassium:proton antiporter activity / sodium:proton antiporter activity / early phagosome / sodium ion import across plasma membrane / potassium ion transmembrane transport / proton transmembrane transport / sodium ion transmembrane transport / regulation of intracellular pH / recycling endosome / phagocytic vesicle membrane / recycling endosome membrane / late endosome membrane / early endosome membrane / defense response to bacterium / protein homodimerization activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
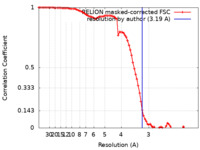
| Method | single particle reconstruction / cryo EM / Resolution: 3.19 Å | |||||||||
 Authors Authors | Winkelmann I / Matsuoka R | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2020 Journal: EMBO J / Year: 2020Title: Structure and elevator mechanism of the mammalian sodium/proton exchanger NHE9. Authors: Iven Winklemann / Rei Matsuoka / Pascal F Meier / Denis Shutin / Chenou Zhang / Laura Orellana / Ricky Sexton / Michael Landreh / Carol V Robinson / Oliver Beckstein / David Drew /    Abstract: Na /H exchangers (NHEs) are ancient membrane-bound nanomachines that work to regulate intracellular pH, sodium levels and cell volume. NHE activities contribute to the control of the cell cycle, cell ...Na /H exchangers (NHEs) are ancient membrane-bound nanomachines that work to regulate intracellular pH, sodium levels and cell volume. NHE activities contribute to the control of the cell cycle, cell proliferation, cell migration and vesicle trafficking. NHE dysfunction has been linked to many diseases, and they are targets of pharmaceutical drugs. Despite their fundamental importance to cell homeostasis and human physiology, structural information for the mammalian NHE was lacking. Here, we report the cryogenic electron microscopy structure of NHE isoform 9 (SLC9A9) from Equus caballus at 3.2 Å resolution, an endosomal isoform highly expressed in the brain and associated with autism spectrum (ASD) and attention deficit hyperactivity (ADHD) disorders. Despite low sequence identity, the NHE9 architecture and ion-binding site are remarkably similar to distantly related bacterial Na /H antiporters with 13 transmembrane segments. Collectively, we reveal the conserved architecture of the NHE ion-binding site, their elevator-like structural transitions, the functional implications of autism disease mutations and the role of phosphoinositide lipids to promote homodimerization that, together, have important physiological ramifications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11067.map.gz emd_11067.map.gz | 47.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11067-v30.xml emd-11067-v30.xml emd-11067.xml emd-11067.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11067_fsc.xml emd_11067_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_11067.png emd_11067.png | 231.1 KB | ||
| Masks |  emd_11067_msk_1.map emd_11067_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11067.cif.gz emd-11067.cif.gz | 7 KB | ||
| Others |  emd_11067_half_map_1.map.gz emd_11067_half_map_1.map.gz emd_11067_half_map_2.map.gz emd_11067_half_map_2.map.gz | 1.5 MB 1.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11067 http://ftp.pdbj.org/pub/emdb/structures/EMD-11067 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11067 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11067 | HTTPS FTP |
-Related structure data
| Related structure data |  6z3zMC  6z3yC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11067.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11067.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0375 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
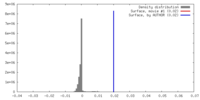
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11067_msk_1.map emd_11067_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_11067_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_11067_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Sodium proton antiporter
| Entire | Name: Sodium proton antiporter |
|---|---|
| Components |
|
-Supramolecule #1: Sodium proton antiporter
| Supramolecule | Name: Sodium proton antiporter / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Sodium/hydrogen exchanger
| Macromolecule | Name: Sodium/hydrogen exchanger / type: protein_or_peptide / ID: 1 / Details: 'GENLYFQ' in C terminal comes from Tag / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.572227 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAVELLVFNF LLILTILTIW LFKNHRFRFL HETGGAMVYG LIMGLILRYA TAPTDIESGT VYDCGKLAFS PSTLLINITD QVYEYKYKR EISQHNINPH LGNAILEKMT FDPEIFFNVL LPPIIFHAGY SLKKRHFFQN LGSILTYAFL GTAISCIVIG L IMYGFVKA ...String: GAVELLVFNF LLILTILTIW LFKNHRFRFL HETGGAMVYG LIMGLILRYA TAPTDIESGT VYDCGKLAFS PSTLLINITD QVYEYKYKR EISQHNINPH LGNAILEKMT FDPEIFFNVL LPPIIFHAGY SLKKRHFFQN LGSILTYAFL GTAISCIVIG L IMYGFVKA MVYAGQLKNG DFHFTDCLFF GSLMSATDPV TVLAIFHELH VDPDLYTLLF GESVLNDAVA IVLTYSISIY SP KENPNAF DAAAFFQSVG NFLGIFAGSF AMGSAYAVVT ALLTKFTKLC EFPMLETGLF FLLSWSAFLS AEAAGLTGIV AVL FCGVTQ AHYTYNNLSL DSKMRTKQLF EFMNFLAENV IFCYMGLALF TFQNHIFNAL FILGAFLAIF VARACNIYPL SFLL NLGRK HKIPWNFQHM MMFSGLRGAI AFALAIRDTE SQPKQMMFST TLLLVFFTVW VFGGGTTGEN LYFQ UniProtKB: Sodium/hydrogen exchanger 9 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 | ||||||||||
| Vitrification | Cryogen name: HELIUM |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Phase plate: ZERNIKE PHASE PLATE / Spherical aberration corrector: Nothing / Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-40 / Average exposure time: 10.0 sec. / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)