[English] 日本語
 Yorodumi
Yorodumi- EMDB-10996: Structure of Mycobacterium smegmatis HelD protein in complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10996 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State I, primary channel engaged | |||||||||
 Map data Map data | Structure of Mycobacterium smegmatis RNA polymerase core in complex with HelD protein (State I) halfmap 2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription cycle helicase-like protein RNA polymerase / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationrecombinational repair / 3'-5' DNA helicase activity / DNA-directed RNA polymerase complex / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / hydrolase activity / protein dimerization activity / response to antibiotic / DNA-templated transcription ...recombinational repair / 3'-5' DNA helicase activity / DNA-directed RNA polymerase complex / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / hydrolase activity / protein dimerization activity / response to antibiotic / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / ATP binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
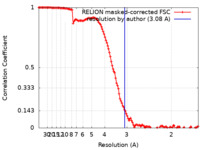
| Method | single particle reconstruction / cryo EM / Resolution: 3.08 Å | |||||||||
 Authors Authors | Kouba T / Koval T | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Mycobacterial HelD is a nucleic acids-clearing factor for RNA polymerase. Authors: Tomáš Kouba / Tomáš Koval' / Petra Sudzinová / Jiří Pospíšil / Barbora Brezovská / Jarmila Hnilicová / Hana Šanderová / Martina Janoušková / Michaela Šiková / Petr Halada / ...Authors: Tomáš Kouba / Tomáš Koval' / Petra Sudzinová / Jiří Pospíšil / Barbora Brezovská / Jarmila Hnilicová / Hana Šanderová / Martina Janoušková / Michaela Šiková / Petr Halada / Michal Sýkora / Ivan Barvík / Jiří Nováček / Mária Trundová / Jarmila Dušková / Tereza Skálová / URee Chon / Katsuhiko S Murakami / Jan Dohnálek / Libor Krásný /    Abstract: RNA synthesis is central to life, and RNA polymerase (RNAP) depends on accessory factors for recovery from stalled states and adaptation to environmental changes. Here, we investigated the mechanism ...RNA synthesis is central to life, and RNA polymerase (RNAP) depends on accessory factors for recovery from stalled states and adaptation to environmental changes. Here, we investigated the mechanism by which a helicase-like factor HelD recycles RNAP. We report a cryo-EM structure of a complex between the Mycobacterium smegmatis RNAP and HelD. The crescent-shaped HelD simultaneously penetrates deep into two RNAP channels that are responsible for nucleic acids binding and substrate delivery to the active site, thereby locking RNAP in an inactive state. We show that HelD prevents non-specific interactions between RNAP and DNA and dissociates stalled transcription elongation complexes. The liberated RNAP can either stay dormant, sequestered by HelD, or upon HelD release, restart transcription. Our results provide insights into the architecture and regulation of the highly medically-relevant mycobacterial transcription machinery and define HelD as a clearing factor that releases RNAP from nonfunctional complexes with nucleic acids. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10996.map.gz emd_10996.map.gz | 84.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10996-v30.xml emd-10996-v30.xml emd-10996.xml emd-10996.xml | 27.5 KB 27.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10996_fsc.xml emd_10996_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10996.png emd_10996.png | 115 KB | ||
| Filedesc metadata |  emd-10996.cif.gz emd-10996.cif.gz | 8.5 KB | ||
| Others |  emd_10996_additional_1.map.gz emd_10996_additional_1.map.gz emd_10996_half_map_1.map.gz emd_10996_half_map_1.map.gz emd_10996_half_map_2.map.gz emd_10996_half_map_2.map.gz | 166.7 MB 141 MB 141 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10996 http://ftp.pdbj.org/pub/emdb/structures/EMD-10996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10996 | HTTPS FTP |
-Validation report
| Summary document |  emd_10996_validation.pdf.gz emd_10996_validation.pdf.gz | 899 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10996_full_validation.pdf.gz emd_10996_full_validation.pdf.gz | 898.6 KB | Display | |
| Data in XML |  emd_10996_validation.xml.gz emd_10996_validation.xml.gz | 19.9 KB | Display | |
| Data in CIF |  emd_10996_validation.cif.gz emd_10996_validation.cif.gz | 25.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10996 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10996 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10996 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10996 | HTTPS FTP |
-Related structure data
| Related structure data |  6yxuMC  6vsxC  6yysC  6z11C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10996.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10996.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Mycobacterium smegmatis RNA polymerase core in complex with HelD protein (State I) halfmap 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8311 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Structure of Mycobacterium smegmatis RNA polymerase core in...
| File | emd_10996_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Mycobacterium smegmatis RNA polymerase core in complex with HelD protein (State I) | ||||||||||||
| Projections & Slices |
| ||||||||||||
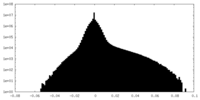
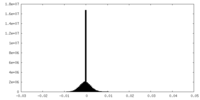
| Density Histograms |
-Half map: Structure of Mycobacterium smegmatis RNA polymerase core in...
| File | emd_10996_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Mycobacterium smegmatis RNA polymerase core in complex with HelD protein (State I) halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
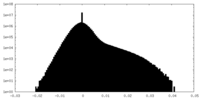
| Density Histograms |
-Half map: Structure of Mycobacterium smegmatis RNA polymerase core in...
| File | emd_10996_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Mycobacterium smegmatis RNA polymerase core in complex with HelD protein (State I) halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
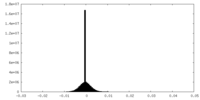
| Density Histograms |
- Sample components
Sample components
-Entire : Mycobacterium smegmatis HelD protein in complex with RNA polymera...
| Entire | Name: Mycobacterium smegmatis HelD protein in complex with RNA polymerase core |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterium smegmatis HelD protein in complex with RNA polymera...
| Supramolecule | Name: Mycobacterium smegmatis HelD protein in complex with RNA polymerase core type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 444 KDa |
-Macromolecule #1: DNA-directed RNA polymerase subunit alpha
| Macromolecule | Name: DNA-directed RNA polymerase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 37.959441 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLISQRPTLS EETVAENRSR FVIEPLEPGF GYTLGNSLRR TLLSSIPGAA VTSIRIDGVL HEFTTVPGVK EDVTDIILNL KGLVVSSDD DEPVTMYLRK QGPGVVTAGD IVPPAGVTVH NPDMHIATLN DKGKLEVELV VERGRGYVPA VQNKASGAEI G RIPVDSIY ...String: MLISQRPTLS EETVAENRSR FVIEPLEPGF GYTLGNSLRR TLLSSIPGAA VTSIRIDGVL HEFTTVPGVK EDVTDIILNL KGLVVSSDD DEPVTMYLRK QGPGVVTAGD IVPPAGVTVH NPDMHIATLN DKGKLEVELV VERGRGYVPA VQNKASGAEI G RIPVDSIY SPVLKVTYKV EATRVEQRTD FDKLIIDVET KNSISPRDAL ASAGGTLVEL FGLARELNAD SEHIEIGPSP AE ADHIASF ALPIDDLDLT VRSYNCLKRE GVHTVGELVA RTESDLLDIR NFGQKSIDEV KIKLHQLGLS LKDSPATFDP SEV AGYDAA TGTWTSDAGY DLDDNQDYAE TEQL UniProtKB: DNA-directed RNA polymerase subunit alpha |
-Macromolecule #2: DNA-directed RNA polymerase subunit beta
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 128.680141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLEGCILAVS SQSKSNAITN NSVPGAPNRV SFAKLREPLE VPGLLDVQTD SFEWLVGSDR WRQAAIDRGE ENPVGGLEEV LAELSPIED FSGSMSLSFS DPRFDEVKAS VDECKDKDMT YAAPLFVTAE FINNNTGEIK SQTVFMGDFP MMTEKGTFII N GTERVVVS ...String: MLEGCILAVS SQSKSNAITN NSVPGAPNRV SFAKLREPLE VPGLLDVQTD SFEWLVGSDR WRQAAIDRGE ENPVGGLEEV LAELSPIED FSGSMSLSFS DPRFDEVKAS VDECKDKDMT YAAPLFVTAE FINNNTGEIK SQTVFMGDFP MMTEKGTFII N GTERVVVS QLVRSPGVYF DETIDKSTEK TLHSVKVIPG RGAWLEFDVD KRDTVGVRID RKRRQPVTVL LKALGWTNEQ IV ERFGFSE IMMGTLEKDT TSGTDEALLD IYRKLRPGEP PTKESAQTLL ENLFFKEKRY DLARVGRYKV NKKLGLNAGK PIT SSTLTE EDVVATIEYL VRLHEGQTSM TVPGGVEVPV EVDDIDHFGN RRLRTVGELI QNQIRVGLSR MERVVRERMT TQDV EAITP QTLINIRPVV AAIKEFFGTS QLSQFMDQNN PLSGLTHKRR LSALGPGGLS RERAGLEVRD VHPSHYGRMC PIETP EGPN IGLIGSLSVY ARVNPFGFIE TPYRKVENGV VTDQIDYLTA DEEDRHVVAQ ANSPTDENGR FTEDRVMVRK KGGEVE FVS ADQVDYMDVS PRQMVSVATA MIPFLEHDDA NRALMGANMQ RQAVPLVRSE APLVGTGMEL RAAIDAGDVV VADKTGV IE EVSADYITVM ADDGTRQSYR LRKFARSNHG TCANQRPIVD AGQRVEAGQV IADGPCTQNG EMALGKNLLV AIMPWEGH N YEDAIILSNR LVEEDVLTSI HIEEHEIDAR DTKLGAEEIT RDIPNVSDEV LADLDERGIV RIGAEVRDGD ILVGKVTPK GETELTPEER LLRAIFGEKA REVRDTSLKV PHGESGKVIG IRVFSREDDD ELPAGVNELV RVYVAQKRKI SDGDKLAGRH GNKGVIGKI LPVEDMPFLP DGTPVDIILN THGVPRRMNI GQILETHLGW VAKAGWNIDV AAGVPDWASK LPEELYSAPA D STVATPVF DGAQEGELAG LLGSTLPNRD GEVMVDADGK STLFDGRSGE PFPYPVTVGY MYILKLHHLV DDKIHARSTG PY SMITQQP LGGKAQFGGQ RFGEMECWAM QAYGAAYTLQ ELLTIKSDDT VGRVKVYEAI VKGENIPEPG IPESFKVLLK ELQ SLCLNV EVLSSDGAAI EMRDGDDEDL ERAAANLGIN LSRNESASVE DLA UniProtKB: DNA-directed RNA polymerase subunit beta |
-Macromolecule #3: DNA-directed RNA polymerase subunit beta'
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta' / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 146.712891 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLDVNFFDEL RIGLATADDI RNWSYGEVKK PETINYRTLK PEKDGLFCEK IFGPTRDWEC YCGKYKRVRF KGIICERCGV EVTRAKVRR ERMGHIELAA PVTHIWYFKG VPSRLGYLLD LAPKDLEKII YFAAYVITSV DDEMRHNELS TLEAEMAVEK K AVEDQRDA ...String: MLDVNFFDEL RIGLATADDI RNWSYGEVKK PETINYRTLK PEKDGLFCEK IFGPTRDWEC YCGKYKRVRF KGIICERCGV EVTRAKVRR ERMGHIELAA PVTHIWYFKG VPSRLGYLLD LAPKDLEKII YFAAYVITSV DDEMRHNELS TLEAEMAVEK K AVEDQRDA DLEARAQKLE ADLAELEAEG AKSDVRRKVR DSGEREMRQL RDRAQRELDR LDEIWNTFTK LAPKQLIVDE VL YRELQDR YGEYFTGAMG AESIKKLIEN FDIDAEAESL REVIRSGKGQ KKLRALKRLK VVAAFQQSGN SPMGMVLDAV PVI PPELRP MVQLDGGRFA TSDLNDLYRR VINRNNRLKR LIDLGAPEII VNNEKRMLQE SVDALFDNGR RGRPVTGPGN RPLK SLSDL LKGKQGRFRQ NLLGKRVDYS GRSVIVVGPQ LKLHQCGLPK LMALELFKPF VMKRLVDLNH AQNIKSAKRM VERQR PQVW DVLEEVIAEH PVLLNRAPTL HRLGIQAFEP QLVEGKAIQL HPLVCEAFNA DFDGDQMAVH LPLSAEAQAE ARILML SSN NILSPASGKP LAMPRLDMVT GLYYLTTLVE GATGEYQAAT KDAPEQGVYS SPAEAIMAMD RGALSVRAKI KVRLTEL RP PTDLEAQLFE NGWKPGDAWT AETTLGRVMF NELLPKSYPF VNEQMHKKVQ ARIINDLAER FPMIVVAQTV DKLKDAGF Y WATRSGVTVS MADVLVPPQK QEILERHEAE ADAIERKYQR GALNHTERNE SLVKIWQDAT EEVGKALEEF YPADNPIIT IVKSGATGNL TQTRTLAGMK GLVTNPKGEF IPRPIKSSFR EGLTVLEYFI NTHGARKGLA DTALRTADSG YLTRRLVDVS QDVIVREHD CETERGINVT LAERGPDGTL IRDAHVETSA FARTLATDAV DANGNVIIER GHDLGDPAID ALLAAGITTV K VRSVLTCT SATGVCAMCY GRSMATGKLV DIGEAVGIVA AQSIGEPGTQ LTMRTFHQGG VTGGADIVGG LPRVQELFEA RV PRNKAPI ADVAGRVRLE ESDKFFKITI VPDDGGEEVV YDKLSKRQRL RVITHEDGTE GVLSDGDHVE VGDQLMEGAA DPH EVLRVQ GPREVQIHLV KEVQEVYRAQ GVSIHDKHIE VIVRQMLRRV TIIDSGSTEF LPGSLTERAE FEAENRRVVA EGGE PAAGR PVLMGITKAS LATDSWLSAA SFQETTRVLT DAAINCRSDK LNGLKENVII GKLIPAGTGI SRYRNIQVQP TEEAR AAAY TIPSYEDQYY SPDFGQATGA AVPLDDYGYS DYR UniProtKB: DNA-directed RNA polymerase subunit beta' |
-Macromolecule #4: DNA-directed RNA polymerase subunit omega
| Macromolecule | Name: DNA-directed RNA polymerase subunit omega / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 11.544763 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTPHADAQL NAADDLGIDS SAASAYDTPL GITNPPIDEL LSRASSKYAL VIYAAKRARQ INDYYNQLGD GILEYVGPLV EPGLQEKPL SIALREIHGD LLEHTEGE UniProtKB: DNA-directed RNA polymerase subunit omega |
-Macromolecule #5: RNA polymerase-associated transcription factor HelD
| Macromolecule | Name: RNA polymerase-associated transcription factor HelD / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 81.298078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRDYEDEL QSERDYVAGL YARLDAERAQ SQRRYAAALR EHGGTAVERD AEVRALAKDI ARLNVADNGL CFGRLDTLDD ARLYIGRLG IFDRDNDFEP LLLDWRAPMA RPFYVATAAN PENMRRRRQF HTLGRKVVDF TDEILGRPTG AEHDATNDAA L LAAVNAPR ...String: MSGRDYEDEL QSERDYVAGL YARLDAERAQ SQRRYAAALR EHGGTAVERD AEVRALAKDI ARLNVADNGL CFGRLDTLDD ARLYIGRLG IFDRDNDFEP LLLDWRAPMA RPFYVATAAN PENMRRRRQF HTLGRKVVDF TDEILGRPTG AEHDATNDAA L LAAVNAPR GEGMRDIVAT IQAEQDQVIR LDHTGVLVIE GGPGTGKTVV ALHRVAYLLY TYRKQMERHG VLVVGPTPAF LD HIGRVLP SLGESDAVFM TPGDFVPGLH VTAEDTPEAA EVKGSLKILD VLKAAVADRQ ELPSEPIPID LSDVTMRIDA ETA KWARDE ARKTGLPHNE ARAEFVDVVT YVVTERAVAR IGRGWLTRDD KHAWEKMRAD VVGELEDHEQ FNAALDALWP ILTP EDVLA QLYTSHERLR AAGAPECLWR ADGEAWTVSD VPLLDELVDL LGRNKAADEA AERERREEEA YAAGVLDLMV DREDL MDDE DHLLAQDLID AEELADRFKE QDNRELSERA AADREWTYGH VVVDEAQELS EMDWRLLMRR CPRRSFTIVG DLAQRR SPA GARSWGAMLD SYVPGRWVYK SLSVNYRTPA EIMAVAAAVL AEFAPDATPP DSVRACGVAP WARQVTDDDI ASAIAEF VS EEAGREGTSV VIGPPDVPGT VPPSETKGLE FDAVLVVEPE RILADGPRGA AELYVALTRA TQRLGVLYRD ALPQALAG L AEGDAAATVE QRTSA UniProtKB: Superfamily protein I DNA or RNA helicase |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Protocol: AB INITIO MODEL | ||||||
| Output model |  PDB-6yxu: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)