+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10889 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vip3Bc1 tetramer in processed, activated state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Toxin / Vip3 / Bt toxin | |||||||||
| Function / homology | Vegetative insecticide protein 3 / Vegetative insecticide protein 3A N terminal / Vegetative insecticidal protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
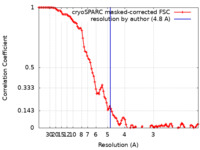
| Method | single particle reconstruction / cryo EM / Resolution: 4.8 Å | |||||||||
 Authors Authors | Thompson RF / Byrne MJ | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Cryo-EM structures of an insecticidal Bt toxin reveal its mechanism of action on the membrane. Authors: Matthew J Byrne / Matthew G Iadanza / Marcos Arribas Perez / Daniel P Maskell / Rachel M George / Emma L Hesketh / Paul A Beales / Marc D Zack / Colin Berry / Rebecca F Thompson /   Abstract: Insect pests are a major cause of crop losses worldwide, with an estimated economic cost of $470 billion annually. Biotechnological tools have been introduced to control such insects without the need ...Insect pests are a major cause of crop losses worldwide, with an estimated economic cost of $470 billion annually. Biotechnological tools have been introduced to control such insects without the need for chemical pesticides; for instance, the development of transgenic plants harbouring genes encoding insecticidal proteins. The Vip3 (vegetative insecticidal protein 3) family proteins from Bacillus thuringiensis convey toxicity to species within the Lepidoptera, and have wide potential applications in commercial agriculture. Vip3 proteins are proposed to exert their insecticidal activity through pore formation, though to date there is no mechanistic description of how this occurs on the membrane. Here we present cryo-EM structures of a Vip3 family toxin in both inactive and activated forms in conjunction with structural and functional data on toxin-membrane interactions. Together these data demonstrate that activated Vip3Bc1 complex is able to insert into membranes in a highly efficient manner, indicating that receptor binding is the likely driver of Vip3 specificity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10889.map.gz emd_10889.map.gz | 96.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10889-v30.xml emd-10889-v30.xml emd-10889.xml emd-10889.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10889_fsc.xml emd_10889_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_10889.png emd_10889.png | 131.5 KB | ||
| Filedesc metadata |  emd-10889.cif.gz emd-10889.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10889 http://ftp.pdbj.org/pub/emdb/structures/EMD-10889 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10889 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10889 | HTTPS FTP |
-Related structure data
| Related structure data |  6yrgMC  7ntxMC  6yrfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10889.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10889.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tetrameric assembly of activated, processed Vip3Bc1
| Entire | Name: Tetrameric assembly of activated, processed Vip3Bc1 |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric assembly of activated, processed Vip3Bc1
| Supramolecule | Name: Tetrameric assembly of activated, processed Vip3Bc1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Vegetative insecticidal protein
| Macromolecule | Name: Vegetative insecticidal protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 91.287148 KDa |
| Recombinant expression | Organism:  Pseudomonas fluorescens (bacteria) Pseudomonas fluorescens (bacteria) |
| Sequence | String: MVQKWMQRMI IVDNNKLNVR ALPSFIDYFN GIYGFATGIK DIMGMIFKTD TGGSNLTLDE ILKNQNLLND ISGKLDGING DLGDLIAQG NLNSELAKEL LKISNEQNQM LNHVNAQLNA INSTLNIYLP KITSMLNEVM KQNHVLSLQI EFLSKQLQEI S DKLDIINL ...String: MVQKWMQRMI IVDNNKLNVR ALPSFIDYFN GIYGFATGIK DIMGMIFKTD TGGSNLTLDE ILKNQNLLND ISGKLDGING DLGDLIAQG NLNSELAKEL LKISNEQNQM LNHVNAQLNA INSTLNIYLP KITSMLNEVM KQNHVLSLQI EFLSKQLQEI S DKLDIINL NVLINSTLTE ITPAYQRIKY VNEKFDELTS TVEKNPKSYQ DNVTKEVIEN LNELTELAKS VTKNDMDSFE FY LQTFHDV MTGNNLFGRS ALKTASELIT KENVTTRGSE IGKVYNFLIV LTSLQAKAFL TLTACRKLLG LTDIDYTQIM NHH IDGQKR EFRINILPTL SNNFSNPSYS KNRGSDIDDP IVVLEAAPGY ALIGFEILND PLPILKGYQA RLKPNYQVDR ESMS ETIYG DIHKLFCPKQ LEQKYYIKDI EFPEGYVITK IVFEKRLNQL GYEVTANFYD PSTGSIDLNK VKVESWKEKS CEEDS CEDE YSIIKAETDG IYMPLGVVSE TFLTPIYGFG LTVDEKNQKI TLTGKSYLRE SLLETDLLNN ETYLIASPDG YISSIV ENW NITSDNTGSW RANNNNAFVD KADTIKGSSS LYTHKDGEFS QFIGNKLKPK TNYVIQYVIK GRPAIYLKNN KDTLFED TK NNFSDFQTVT KKFNSGVNPS EIYFLFKNQS EYEAWGNNFI ILEIKSLEFL PQMLKPEDWI PSGNVQMKDG GRLEILGD G YFKQFIKLEN DSTYHLRLSV KGTGRVSIID ESKYLLFVNV KDEDLTRVIK NTSSKGECFI ALEGTYVENS STIFSNVSI VKE UniProtKB: Vegetative insecticidal protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: FEI FALCON III (4k x 4k) / #0 - Detector mode: INTEGRATING / #0 - Number grids imaged: 1 / #0 - Average exposure time: 1.5 sec. / #0 - Average electron dose: 70.8 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: FEI FALCON III (4k x 4k) / #1 - Detector mode: INTEGRATING / #1 - Average electron dose: 42.51 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)