+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10843 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mono-ubiquitinated FANCD2 in complex with FANCI | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
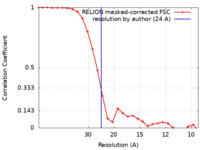
| Method | single particle reconstruction / cryo EM / Resolution: 24.0 Å | |||||||||
 Authors Authors | Rennie ML / Lemonidis K / Arkinson C / Chaugule VK / Clarke M / Streetley J / Spagnolo L / Walden H | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2020 Journal: EMBO Rep / Year: 2020Title: Differential functions of FANCI and FANCD2 ubiquitination stabilize ID2 complex on DNA. Authors: Martin L Rennie / Kimon Lemonidis / Connor Arkinson / Viduth K Chaugule / Mairi Clarke / James Streetley / Laura Spagnolo / Helen Walden /  Abstract: The Fanconi anaemia (FA) pathway is a dedicated pathway for the repair of DNA interstrand crosslinks and is additionally activated in response to other forms of replication stress. A key step in the ...The Fanconi anaemia (FA) pathway is a dedicated pathway for the repair of DNA interstrand crosslinks and is additionally activated in response to other forms of replication stress. A key step in the FA pathway is the monoubiquitination of each of the two subunits (FANCI and FANCD2) of the ID2 complex on specific lysine residues. However, the molecular function of these modifications has been unknown for nearly two decades. Here, we find that ubiquitination of FANCD2 acts to increase ID2's affinity for double-stranded DNA via promoting a large-scale conformational change in the complex. The resulting complex encircles DNA, by forming a secondary "Arm" ID2 interface. Ubiquitination of FANCI, on the other hand, largely protects the ubiquitin on FANCD2 from USP1-UAF1 deubiquitination, with key hydrophobic residues of FANCI's ubiquitin being important for this protection. In effect, both of these post-translational modifications function to stabilize a conformation in which the ID2 complex encircles DNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10843.map.gz emd_10843.map.gz | 271.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10843-v30.xml emd-10843-v30.xml emd-10843.xml emd-10843.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10843_fsc.xml emd_10843_fsc.xml | 2.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_10843.png emd_10843.png | 30.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10843 http://ftp.pdbj.org/pub/emdb/structures/EMD-10843 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10843 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10843 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10843.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10843.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.784 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Binary complex of mono-ubiquitinated FANCD2 and FANCI
| Entire | Name: Binary complex of mono-ubiquitinated FANCD2 and FANCI |
|---|---|
| Components |
|
-Supramolecule #1: Binary complex of mono-ubiquitinated FANCD2 and FANCI
| Supramolecule | Name: Binary complex of mono-ubiquitinated FANCD2 and FANCI / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 330 KDa |
-Macromolecule #1: FANCD2 mono-ubiquitinated at K561
| Macromolecule | Name: FANCD2 mono-ubiquitinated at K561 / type: protein_or_peptide / ID: 1 Details: Additional single ubiquitin conjugated to K561 (ubiquitin sequence: GPGSMQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLSDYNIQKESTLHLVLRLRGG) Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: GPGSMVSKRR LSKSEDKESL TEDASKTRKQ PLSKKTKKSH IANEVEENDS IFVKLL KIS GIILKTGESQ NQLAVDQIAF QKKLFQTL R RHPSYPKIIE EFVSGLESYI EDEDSFRNCL LSCERLQDE EASMGASYSK SLIKLLLGID ILQPAIIKTL FEKLPEYFFE ...String: GPGSMVSKRR LSKSEDKESL TEDASKTRKQ PLSKKTKKSH IANEVEENDS IFVKLL KIS GIILKTGESQ NQLAVDQIAF QKKLFQTL R RHPSYPKIIE EFVSGLESYI EDEDSFRNCL LSCERLQDE EASMGASYSK SLIKLLLGID ILQPAIIKTL FEKLPEYFFE NKNSDEINIP RLIVSQLKWL DRVVDGKDL TTKI MQLIS IAPENLQHDI ITSLPEILGD SQHADVGKEL SDLLIENTSL TVPILDVLSS LRLDPNFLLK VRQLVMD KL SSIRLEDLPV IIK FILHSV TAMDTLEVIS ELREKLDLQH CVLPSRLQAS QVKLKSKGRA SSSGNQESS GQSCIILLFD VIKSAIRYEK TISEAWIKAI ENTASVSEHK VFDLV MLFI IYSTNTQTKK YIDRVLRNKI RSGCI QEQL LQSTFSVHYL VLKDMCSSIL SLAQSLLHSL DQSIISFGSL LYKYAFKFFD TYCQQEVVGA LVTHICS GN EAEVDTALDV LLELVVLNPS AMMMNAVFVK GILDYLDNIS PQQIRKLFYV LSTLAFSKQN EASSHIQD D MHLVIRKQLS STVFKYKLIG IIGAVTMAG IMAADRSESP SLTQERANLS DEQCTQVTSL LQLVHSCSE QSPQASALYY DEFANLIQHE KLDPKALEWV GHTICNDFQD AFVVDSCVVP EGDFPFPVKA L YGLEEYD TQDGIAINLL PLLFSQDFAK DGGPVTSQES GQKLVSPLCL APYFRLLRLC VERQHNGNLE EIDGLLDCPI FLTDLEPGE KLESMSAKER SFMC SLIFL TLNWFREIVN AFCQETSPEM KGKVLTRLKH IVELQIILEK YLA VTPDYV PPLGNFDVET LDITPHTVTA ISAKIRKKGK IERKQKTDGS KTSSSD TLS EEKNSECDPT PSHRG QLNK EFTGKEEKTS LLLHNSHAFF RELDIEVFSI LHCGLVTKFI LDTEMHTEAT EVVQLGPPEL LFLLEDL SQ KLESMLTPP IARRVPFLKN KGSRNIGFSH LQQRSAQEIV HCVFQLLTPM CNHLENIHNY FQCLAAEN H GVVDGPGVKV QEYHIMSSCY QRLLQIFHGL FAWSGFSQPE N QNLLYSAL HVLSSRLKQG EHSQPLE EL LSQSVHYLQN FHQSIPSFQC ALYLIRLLMV ILEKSTASAQ NKEKIASLAR QFLCRVWPSG DKEKSNIS N DQLH ALLCI YLEHTESILK AIEEIAGVGV PELINSPKDA SSSTFPTLTR HTFVVFFRVM MAELEKTVKK IE PGTAADS QQIHEEKLLY WNMAVRDFSI LINLIK VFD SHPVLHVCLK YGRLFVEAFL KQCMPLLDFS FRK HREDVL SLLETFQLDT RLLHHLCGHS KIHQDTRLTQ HVPLLKKTLE LLVCRVKAML TLNNCREA F WLGN LKNRD LQGEEIKSQN SQESTADESE DDMSSQASKS KATEDGEEDE VSAGEKEQDS DESYDDSD |
-Macromolecule #2: FANCI
| Macromolecule | Name: FANCI / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHENL YFQGKPIPNP LLGLDSTMDQ KILSLAAEKT ADKLQEFLQT LREGDLTNLL QNQAVKGKV AGALLRAIFK GSPCSEEAGT LRRRKIYTCC I QLVESGDL QKEIASEIIG LLMLEAHHFP GPLLVELANE FIS AVREGS LVNGKSLELL PIILTALATK ...String: MHHHHHHENL YFQGKPIPNP LLGLDSTMDQ KILSLAAEKT ADKLQEFLQT LREGDLTNLL QNQAVKGKV AGALLRAIFK GSPCSEEAGT LRRRKIYTCC I QLVESGDL QKEIASEIIG LLMLEAHHFP GPLLVELANE FIS AVREGS LVNGKSLELL PIILTALATK KENLAYGKGV LSGEECKKQL INTLCSGRWD QQY VIQLTS MFKD VPLTA EEVEFVVEKA LSMFSKMNLQ EIPPLVYQLL VLSSKGSRKS VLEGIIAFFS ALDKQHNEEQ SGDEL LDVV TVPSGELRHV EGTIIL HIV FAIKLDYELG RELVKHLKVG QQGDSNNNLS PFSIALLLSV TRIQRFQD Q VLDLLKTSVV KSFKDLQLLQ GSKFLQNLVP HRSYVSTMIL EVVKNSVH S WDHVTQGLVE LGFILMDS Y GPKKVLDGKT IETSPSLSRM PNQHACKLGA NILLETFKIH EMIRQEILEQ VLNRVVTRAS SPISHFLDLL SNIVMYAPL V LQSCSSKV TEAFDYLSFL PLQTVQRLLK AVQPLLKVSM SMRDCLILVL RKAMFANQLD A RKSAVAGF LLLLKNFKVL GSLSSSQCSQ SLSVSQVHVD VHS HYNSVA NETFCLEIMD SLRRCLSQQA D VRLMLYEG FYDVLRRNSQ LANSVMQTLL SQLKQFYEPK PDLLPPLKLE ACILTQGDKI SLQEPLDYLL CC IQH CLA WYKNTVIPLQ QGEEEEEEEE AFYEDLDDIL ESITNRMIKS ELEDFELDKS ADFSQSTSIG IKNNI CAFL VMGVCEVLIE YNFSISSFSK NRFEDILS L FMCYKKLSDI LNEKAGKAKT KMANKTSDSL LSMKFVS SL LTALFRDSIQ SHQESLSVLR SSNEFMRYAV NVALQKVQQL KETGHVSGPD GQNPEKIFQN LCDITR VL LWRYTSIPTS VEESGKKEKG KSISLLCLEG LQKIFSAVQQ FYQPKIQQFL RALDVTDKEG EEREDADV S VTQRTAFQIR QFQRSLLNLL SSQ EEDFNS KEALLLVTVL TSLSKLLEPS SPQFVQMLSW TSKICKENS REDALFCKSL MNLLFSLHVS YKSPVILLRD LSQDIHGHLG DIDQDVEVEK TNHFA IVNL RTAAPTVCLL V LSQAEKVL EEVDWLITKL KGQVSQETLS EEASSQATLP NQPVEKAIIM QLGTLLTFFH ELVQTALPSG SC VDTLLKD LCKMYTTL T ALVRYYLQVC QSSGGIPKNM EKLVKLSGSH LTPLCYSFIS YVQNKSKSLN YTG EKKEKP AAVATAMARV LRETKPIPNL IFAIEQYEKF LIHLSKKSKV NLMQHMKLS TSRDFKIKGN ILDMVLREDG EDENEEGTAS EHGGQNKEPA KKKRKK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.55 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | Complex components were mixed then sample was exchanged into cryoEM buffer via a desalting column |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-64 (8k x 8k) / Detector mode: COUNTING / Average electron dose: 46.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)