[English] 日本語
 Yorodumi
Yorodumi- EMDB-10792: Hybrid structure of the SPP1 tail tube by solid-state NMR and cry... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10792 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - Final EM Refinement | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / tail tube / scaffolding / DNA transport / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral head-tail joining / virus tail, tube / symbiont genome ejection through host cell envelope, long flexible tail mechanism / viral translational frameshifting Similarity search - Function | |||||||||
| Biological species |  Bacillus phage SPP1 (virus) Bacillus phage SPP1 (virus) | |||||||||
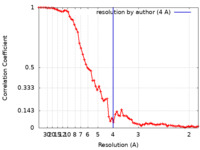
| Method | helical reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Zinke M / Sachowsky KAA | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Architecture of the flexible tail tube of bacteriophage SPP1. Authors: Maximilian Zinke / Katrin A A Sachowsky / Carl Öster / Sophie Zinn-Justin / Raimond Ravelli / Gunnar F Schröder / Michael Habeck / Adam Lange /    Abstract: Bacteriophage SPP1 is a double-stranded DNA virus of the Siphoviridae family that infects the bacterium Bacillus subtilis. This family of phages features a long, flexible, non-contractile tail that ...Bacteriophage SPP1 is a double-stranded DNA virus of the Siphoviridae family that infects the bacterium Bacillus subtilis. This family of phages features a long, flexible, non-contractile tail that has been difficult to characterize structurally. Here, we present the atomic structure of the tail tube of phage SPP1. Our hybrid structure is based on the integration of structural restraints from solid-state nuclear magnetic resonance (NMR) and a density map from cryo-EM. We show that the tail tube protein gp17.1 organizes into hexameric rings that are stacked by flexible linker domains and, thus, form a hollow flexible tube with a negatively charged lumen suitable for the transport of DNA. Additionally, we assess the dynamics of the system by combining relaxation measurements with variances in density maps. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: Spinal Column Architecture of the Flexible SPP1 Bacteriophage Tail Tube Authors: Zinke M / Sachowsky KAA / Oster C / Zinn-Justin S / Ravelli R / Schroder GF / Habeck M / Lange A | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10792.map.gz emd_10792.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10792-v30.xml emd-10792-v30.xml emd-10792.xml emd-10792.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10792_fsc.xml emd_10792_fsc.xml | 7 KB | Display |  FSC data file FSC data file |
| Images |  emd_10792.png emd_10792.png | 95.8 KB | ||
| Others |  emd_10792_half_map_1.map.gz emd_10792_half_map_1.map.gz emd_10792_half_map_2.map.gz emd_10792_half_map_2.map.gz | 27.7 MB 27.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10792 http://ftp.pdbj.org/pub/emdb/structures/EMD-10792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10792 | HTTPS FTP |
-Validation report
| Summary document |  emd_10792_validation.pdf.gz emd_10792_validation.pdf.gz | 814 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10792_full_validation.pdf.gz emd_10792_full_validation.pdf.gz | 813.6 KB | Display | |
| Data in XML |  emd_10792_validation.xml.gz emd_10792_validation.xml.gz | 12 KB | Display | |
| Data in CIF |  emd_10792_validation.cif.gz emd_10792_validation.cif.gz | 17.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10792 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10792 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10792 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10792 | HTTPS FTP |
-Related structure data
| Related structure data |  6yegMC  6yq5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10792.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10792.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.935 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_10792_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10792_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SPP1 tail tube
| Entire | Name: SPP1 tail tube |
|---|---|
| Components |
|
-Supramolecule #1: SPP1 tail tube
| Supramolecule | Name: SPP1 tail tube / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Bacillus phage SPP1 (virus) Bacillus phage SPP1 (virus) |
-Macromolecule #1: Tail tube protein gp17.1*
| Macromolecule | Name: Tail tube protein gp17.1* / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacillus phage SPP1 (virus) Bacillus phage SPP1 (virus) |
| Molecular weight | Theoretical: 19.674451 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPIMGQDVKY LFQSIDAATG SAPLFPAYQT DGSVSGEREL FDEQTKNGRI LGPGSVADSG EVTYYGKRGD AGQKAIEDAY QNGKQIKFW RVDTVKNEND KYDAQFGFAY IESREYSDGV EGAVEISISL QVIGELKNGE IDTLPEEIVN VSKGGYDFQQ P GQTTGEAP GTVPAPHHHH HH UniProtKB: Tail tube protein gp17.1* |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 3.0 sec. / Average electron dose: 70.0 e/Å2 Details: Images were collected in movie-mode at 20 fractions, each fraction had 6 frames. Dose rate was 23 e/A^2/s, i.e. dose per frame was 3.5 electrons/A^2 . |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)