[English] 日本語
 Yorodumi
Yorodumi- EMDB-10791: 70S initiation complex with assigned rRNA modifications from Stap... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10791 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 70S initiation complex with assigned rRNA modifications from Staphylococcus aureus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | rRNA modifications / 70S / initiation complex / S. aureus / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationlarge ribosomal subunit / transferase activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit ...large ribosomal subunit / transferase activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / RNA binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Staphylococcus aureus subsp. aureus NCTC 8325 (bacteria) / synthetic construct (others) / Staphylococcus aureus subsp. aureus NCTC 8325 (bacteria) / synthetic construct (others) /   Staphylococcus aureus (strain NCTC 8325) (bacteria) Staphylococcus aureus (strain NCTC 8325) (bacteria) | |||||||||
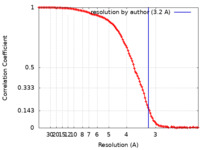
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Fatkhullin B / Golubev A | |||||||||
| Funding support |  Russian Federation, Russian Federation,  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: FEBS Lett / Year: 2020 Journal: FEBS Lett / Year: 2020Title: Cryo-EM structure of the ribosome functional complex of the human pathogen Staphylococcus aureus at 3.2 Å resolution. Authors: Alexander Golubev / Bulat Fatkhullin / Iskander Khusainov / Lasse Jenner / Azat Gabdulkhakov / Shamil Validov / Gulnara Yusupova / Marat Yusupov / Konstantin Usachev /    Abstract: Staphylococcus aureus is a bacterial pathogen and one of the leading causes of healthcare-acquired infections in the world. The growing antibiotic resistance of S. aureus obliges us to search for ...Staphylococcus aureus is a bacterial pathogen and one of the leading causes of healthcare-acquired infections in the world. The growing antibiotic resistance of S. aureus obliges us to search for new drugs and treatments. As the majority of antibiotics target the ribosome, knowledge of its detailed structure is crucial for drug development. Here, we report the cryo-EM reconstruction at 3.2 Å resolution of the S. aureus ribosome with P-site tRNA, messenger RNA, and 10 RNA modification sites previously not assigned or visualized. The resulting model is the most precise and complete high-resolution structure to date of the S. aureus 70S ribosome with functional ligands. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10791.map.gz emd_10791.map.gz | 96.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10791-v30.xml emd-10791-v30.xml emd-10791.xml emd-10791.xml | 77.8 KB 77.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10791_fsc.xml emd_10791_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_10791.png emd_10791.png | 132.2 KB | ||
| Filedesc metadata |  emd-10791.cif.gz emd-10791.cif.gz | 14.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10791 http://ftp.pdbj.org/pub/emdb/structures/EMD-10791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10791 | HTTPS FTP |
-Related structure data
| Related structure data |  6yefMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10791.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10791.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : 70S ribosome
+Supramolecule #1: 70S ribosome
+Supramolecule #2: 16S rRNA, 30S ribosomal subunit
+Supramolecule #3: 23S and 5S rRNA, 50S ribosomal subunit
+Supramolecule #4: mRNA
+Supramolecule #5: E-site tRNA
+Macromolecule #1: 16S rRNA
+Macromolecule #21: mRNA
+Macromolecule #22: 23S rRNA
+Macromolecule #23: 5S rRNA
+Macromolecule #52: P-site tRNA
+Macromolecule #2: 30S ribosomal protein S2
+Macromolecule #3: 30S ribosomal protein S3
+Macromolecule #4: 30S ribosomal protein S4
+Macromolecule #5: 30S ribosomal protein S5
+Macromolecule #6: 30S ribosomal protein S6
+Macromolecule #7: 30S ribosomal protein S7
+Macromolecule #8: 30S ribosomal protein S8
+Macromolecule #9: 30S ribosomal protein S9
+Macromolecule #10: ribosomal protein uS10
+Macromolecule #11: 30S ribosomal protein S11
+Macromolecule #12: 30S ribosomal protein S12
+Macromolecule #13: 30S ribosomal protein S13
+Macromolecule #14: 30S ribosomal protein S14
+Macromolecule #15: 30S ribosomal protein S15
+Macromolecule #16: 30S ribosomal protein S16
+Macromolecule #17: 30S ribosomal protein S17
+Macromolecule #18: 30S ribosomal protein S18
+Macromolecule #19: 30S ribosomal protein S19
+Macromolecule #20: 30S ribosomal protein S20
+Macromolecule #24: 50S ribosomal protein L2
+Macromolecule #25: 50S ribosomal protein L3
+Macromolecule #26: 50S ribosomal protein L4
+Macromolecule #27: 50S ribosomal protein L5
+Macromolecule #28: 50S ribosomal protein L6
+Macromolecule #29: 50S ribosomal protein L13
+Macromolecule #30: 50S ribosomal protein L14
+Macromolecule #31: 50S ribosomal protein L15
+Macromolecule #32: 50S ribosomal protein L16
+Macromolecule #33: 50S ribosomal protein L17
+Macromolecule #34: 50S ribosomal protein L18
+Macromolecule #35: 50S ribosomal protein L19
+Macromolecule #36: 50S ribosomal protein L20
+Macromolecule #37: 50S ribosomal protein L21
+Macromolecule #38: 50S ribosomal protein L22
+Macromolecule #39: 50S ribosomal protein L23
+Macromolecule #40: 50S ribosomal protein L24
+Macromolecule #41: 50S ribosomal protein L25
+Macromolecule #42: 50S ribosomal protein L27
+Macromolecule #43: 50S ribosomal protein L28
+Macromolecule #44: 50S ribosomal protein L29
+Macromolecule #45: 50S ribosomal protein L30
+Macromolecule #46: 50S ribosomal protein L31 type B
+Macromolecule #47: 50S ribosomal protein L32
+Macromolecule #48: 50S ribosomal protein L33 2
+Macromolecule #49: 50S ribosomal protein L34
+Macromolecule #50: 50S ribosomal protein L35
+Macromolecule #51: 50S ribosomal protein L36
+Macromolecule #53: MAGNESIUM ION
+Macromolecule #54: POTASSIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)