[English] 日本語
 Yorodumi
Yorodumi- EMDB-1077: Visualization of ribosome-recycling factor on the Escherichia col... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1077 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. | |||||||||
 Map data Map data | Complex of e.coli 70S ribosome and ribosome recycling factor (RRF). | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | Ribosome recycling factor / Ribosome recycling factor domain / RRF superfamily / Ribosome recycling factor / ribosomal large subunit binding / translational termination / cytoplasm / Ribosome-recycling factor Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 12.0 Å | |||||||||
 Authors Authors | Agrawal RK | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2004 Journal: Proc Natl Acad Sci U S A / Year: 2004Title: Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Authors: Rajendra K Agrawal / Manjuli R Sharma / Michael C Kiel / Go Hirokawa / Timothy M Booth / Christian M T Spahn / Robert A Grassucci / Akira Kaji / Joachim Frank /  Abstract: After the termination step of protein synthesis, a deacylated tRNA and mRNA remain associated with the ribosome. The ribosome-recycling factor (RRF), together with elongation factor G (EF-G), ...After the termination step of protein synthesis, a deacylated tRNA and mRNA remain associated with the ribosome. The ribosome-recycling factor (RRF), together with elongation factor G (EF-G), disassembles this posttermination complex into mRNA, tRNA, and the ribosome. We have obtained a three-dimensional cryo-electron microscopic map of a complex of the Escherichia coli 70S ribosome and RRF. We find that RRF interacts mainly with the segments of the large ribosomal subunit's (50S) rRNA helices that are involved in the formation of two central intersubunit bridges, B2a and B3. The binding of RRF induces considerable conformational changes in some of the functional domains of the ribosome. As compared to its binding position derived previously by hydroxyl radical probing study, we find that RRF binds further inside the intersubunit space of the ribosome such that the tip of its domain I is shifted (by approximately 13 A) toward protein L5 within the central protuberance of the 50S subunit, and domain II is oriented more toward the small ribosomal subunit (30S). Overlapping binding sites of RRF, EF-G, and the P-site tRNA suggest that the binding of EF-G would trigger the removal of deacylated tRNA from the P site by moving RRF toward the ribosomal E site, and subsequent removal of mRNA may be induced by a shift in the position of 16S rRNA helix 44, which harbors part of the mRNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1077.map.gz emd_1077.map.gz | 7.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1077-v30.xml emd-1077-v30.xml emd-1077.xml emd-1077.xml | 10.9 KB 10.9 KB | Display Display |  EMDB header EMDB header |
| Images |  1077.gif 1077.gif | 16.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1077 http://ftp.pdbj.org/pub/emdb/structures/EMD-1077 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1077 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1077 | HTTPS FTP |
-Related structure data
| Related structure data |  1t1mMC  1t1oMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1077.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1077.map.gz / Format: CCP4 / Size: 8.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complex of e.coli 70S ribosome and ribosome recycling factor (RRF). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
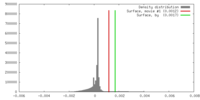
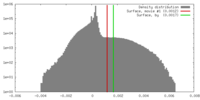
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 70S-RRF complex of e.coli
| Entire | Name: 70S-RRF complex of e.coli |
|---|---|
| Components |
|
-Supramolecule #1000: 70S-RRF complex of e.coli
| Supramolecule | Name: 70S-RRF complex of e.coli / type: sample / ID: 1000 / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 2.5 MDa / Theoretical: 2.5 MDa / Method: sedimentation |
-Supramolecule #1: 70S
| Supramolecule | Name: 70S / type: complex / ID: 1 / Ribosome-details: ribosome-prokaryote: ALL |
|---|---|
| Molecular weight | Experimental: 2.48 MDa / Theoretical: 2.48 MDa |
-Macromolecule #1: RRF
| Macromolecule | Name: RRF / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 20 KDa / Theoretical: 20 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 52 mM Tris-HCL (pH7.5), 25 mM KCL, 5.5 mM NH4Cl, 11 mM Mg(OAc)2 0.3 mM Beta-ME |
|---|---|
| Grid | Details: quantifoil grids with holy carbon |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 30 % / Chamber temperature: 93 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: two side blotting plunger Method: Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 93 K |
| Date | Jun 1, 2002 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 14 µm / Number real images: 76 / Average electron dose: 20 e/Å2 / Od range: 1.2 / Bits/pixel: 12 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 49696 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.4 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Cryo transfer / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: defocus groups |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 12.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER Details: Final map was calculated from 23 CTF corrected defocus groups Number images used: 51217 |
| Final angle assignment | Details: SPIDER: theta 15 degrees, phi 15 degrees |
-Atomic model buiding 1
| Software | Name: manual |
|---|---|
| Details | Protocol: Rigid Body. the RRF was fitted as single rigid body |
| Refinement | Protocol: RIGID BODY FIT / Target criteria: cross correlation coefficient |
| Output model |  PDB-1t1m:  PDB-1t1o: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)