+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10722 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
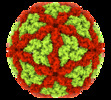
| Title | Capsid structure of Leishmania RNA virus 1 | |||||||||
 Map data Map data | Map rotated to crystallographic orientation with origin in 000. | |||||||||
 Sample Sample | Leishmania RNA virus 1 - 4 != Leishmania RNA virus 1 - LgM5313 Leishmania RNA virus 1 - 4
| |||||||||
 Keywords Keywords | virus-like particle / capsid structure / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Totivirus coat / Totivirus coat protein / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Leishmania RNA virus 1 - 4 / Leishmania RNA virus 1 - 4 /  Leishmania RNA virus 1 - LgM5313 Leishmania RNA virus 1 - LgM5313 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.65 Å | |||||||||
 Authors Authors | Prochazkova M / Fuzik T | |||||||||
| Funding support |  Czech Republic, 1 items Czech Republic, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2021 Journal: J Virol / Year: 2021Title: Capsid Structure of RNA Virus 1. Authors: Michaela Procházková / Tibor Füzik / Danyil Grybchuk / Francesco Luca Falginella / Lucie Podešvová / Vyacheslav Yurchenko / Robert Vácha / Pavel Plevka /   Abstract: parasites cause a variety of symptoms, including mucocutaneous leishmaniasis, which results in the destruction of the mucous membranes of the nose, mouth, and throat. The species of carrying RNA ... parasites cause a variety of symptoms, including mucocutaneous leishmaniasis, which results in the destruction of the mucous membranes of the nose, mouth, and throat. The species of carrying RNA virus 1 (LRV1), from the family , are more likely to cause severe disease and are less sensitive to treatment than those that do not contain the virus. Although the importance of LRV1 for the severity of leishmaniasis was discovered a long time ago, the structure of the virus remained unknown. Here, we present a cryo-electron microscopy reconstruction of the virus-like particle of LRV1 determined to a resolution of 3.65 Å. The capsid has icosahedral symmetry and is formed by 120 copies of a capsid protein assembled in asymmetric dimers. RNA genomes of viruses from the family are synthetized, but not capped at the 5' end, by virus RNA polymerases. To protect viral RNAs from degradation, capsid proteins of the L-A totivirus cleave the 5' caps of host mRNAs, creating decoys to overload the cellular RNA quality control system. Capsid proteins of LRV1 form positively charged clefts, which may be the cleavage sites for the 5' cap of mRNAs. The putative RNA binding site of LRV1 is distinct from that of the related L-A virus. The structure of the LRV1 capsid enables the rational design of compounds targeting the putative decapping site. Such inhibitors may be developed into a treatment for mucocutaneous leishmaniasis caused by LRV1-positive species of Twelve million people worldwide suffer from leishmaniasis, resulting in more than 30 thousand deaths annually. The disease has several variants that differ in their symptoms. The mucocutaneous form, which leads to disintegration of the nasal septum, lips, and palate, is caused predominantly by parasites carrying RNA virus 1 (LRV1). Here, we present the structure of the LRV1 capsid determined using cryo-electron microscopy. Capsid proteins of a related totivirus, L-A virus, protect viral RNAs from degradation by cleaving the 5' caps of host mRNAs. Capsid proteins of LRV1 may have the same function. We show that the LRV1 capsid contains positively charged clefts that may be sites for the cleavage of mRNAs of cells. The structure of the LRV1 capsid enables the rational design of compounds targeting the putative mRNA cleavage site. Such inhibitors may be used as treatments for mucocutaneous leishmaniasis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10722.map.gz emd_10722.map.gz | 95.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10722-v30.xml emd-10722-v30.xml emd-10722.xml emd-10722.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10722_fsc.xml emd_10722_fsc.xml | 19.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_10722.png emd_10722.png | 271.5 KB | ||
| Masks |  emd_10722_msk_1.map emd_10722_msk_1.map | 620.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10722.cif.gz emd-10722.cif.gz | 6.5 KB | ||
| Others |  emd_10722_additional_1.map.gz emd_10722_additional_1.map.gz | 109.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10722 http://ftp.pdbj.org/pub/emdb/structures/EMD-10722 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10722 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10722 | HTTPS FTP |
-Validation report
| Summary document |  emd_10722_validation.pdf.gz emd_10722_validation.pdf.gz | 623.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10722_full_validation.pdf.gz emd_10722_full_validation.pdf.gz | 622.9 KB | Display | |
| Data in XML |  emd_10722_validation.xml.gz emd_10722_validation.xml.gz | 16.9 KB | Display | |
| Data in CIF |  emd_10722_validation.cif.gz emd_10722_validation.cif.gz | 23.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10722 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10722 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10722 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10722 | HTTPS FTP |
-Related structure data
| Related structure data |  6y83MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10722.map.gz / Format: CCP4 / Size: 620.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10722.map.gz / Format: CCP4 / Size: 620.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map rotated to crystallographic orientation with origin in 000. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
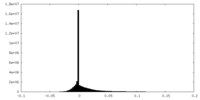
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10722_msk_1.map emd_10722_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
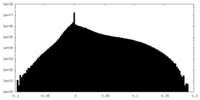
| Density Histograms |
-Additional map: Map in original position, matches mask.
| File | emd_10722_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map in original position, matches mask. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Leishmania RNA virus 1 - 4
| Entire | Name:  Leishmania RNA virus 1 - 4 Leishmania RNA virus 1 - 4 |
|---|---|
| Components |
|
-Supramolecule #1: Leishmania RNA virus 1 - LgM5313
| Supramolecule | Name: Leishmania RNA virus 1 - LgM5313 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1285422 / Sci species name: Leishmania RNA virus 1 - LgM5313 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Leishmania guyanensis (eukaryote) / Strain: MHOM/BR/75/M4147 Leishmania guyanensis (eukaryote) / Strain: MHOM/BR/75/M4147 |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 422.0 Å / T number (triangulation number): 2 |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 Details: Subunit A model for residues 15-204, 210-520, 541-635. Subunit B model for residues 19-290, 300-517, 541-576, 583-642.,Subunit A model for residues 15-204, 210-520, 541-635. Subunit B model ...Details: Subunit A model for residues 15-204, 210-520, 541-635. Subunit B model for residues 19-290, 300-517, 541-576, 583-642.,Subunit A model for residues 15-204, 210-520, 541-635. Subunit B model for residues 19-290, 300-517, 541-576, 583-642. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Leishmania RNA virus 1 - 4 Leishmania RNA virus 1 - 4 |
| Molecular weight | Theoretical: 87.53693 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MADIPNSDKI ACGRKPMFCE IIKLANRKRL IFNTTDERVY DARLNYCSTA DSTVQADCHI YWRLKLRRTD AVFEEYTGQG YSLDTAAYP QQYTDIIRGY YSKHVSSSLA ANTQHFVNVL AMLRHAGACI AHYCMTGKID FDILSKKKHK NKEVVTLSNA D SLSFLPHS ...String: MADIPNSDKI ACGRKPMFCE IIKLANRKRL IFNTTDERVY DARLNYCSTA DSTVQADCHI YWRLKLRRTD AVFEEYTGQG YSLDTAAYP QQYTDIIRGY YSKHVSSSLA ANTQHFVNVL AMLRHAGACI AHYCMTGKID FDILSKKKHK NKEVVTLSNA D SLSFLPHS ALYLPSPLRA SDPEIFNMLY LLGCACDASI AMDNISNTSG AAKYSMPHYN PLQLSHALHV TIFYMLSLMD SC GYGDDAV LALTSGLHSV TTVIAHSDEG GITRDALREL SYTQPYGTMP VPIAGYFQHI NVLFTTQPAW DQFAGIWDYV ILA TAALVH LSDPGMTVND VTYPTTLTTK VATVDGRNSD LAAQMMHSAT RFCDIFVENL STFWGVVANP DGNASQALLH AFNI VACAV EPNRHLEMNV MAPWYWVESS ALFCDYAPFR SPISSAGYGP QCVYGARLVL AATNSLEFTG EAGDYSAYRF EWTTM RHNP LFNILNKRVG DGLANVDFRL RPFNEWLLEG QPSRRSCNSA GHGTPTATCS HKTPNHDTLD EYIWGSTSCD LFHPAE LTS YTAVCVRFRN YLSGADGDVR ILNTPTREVI EGNVVTRCDG IRCLDSNKRI QHVPEVARRY CMMARYLAQA RTFGALT IG DDIIRGFDKV EKIVKMHKSN NRLDQMPLID VTGLCQPMIE TSTVRASTTT RIDPNKLAAA TARVELPLAP RCTSSLIP S SDTVPEPEPQ VGEPGDNGGC APDLGTGGGS GIEGRGSMDI GDPNSVQALA RLQASSVDKL AAALEHHHHH HHH UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK II / Details: blot force 3 blot time 5 wait time 10. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 12000 / Average exposure time: 0.5 sec. / Average electron dose: 21.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.0005 µm / Nominal defocus min: -0.0005 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)