[English] 日本語
 Yorodumi
Yorodumi- EMDB-10658: CryoEM structure of the ring-shaped virulence factor EspB from My... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10658 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the ring-shaped virulence factor EspB from Mycobacterium tuberculosis | |||||||||
 Map data Map data | Sharped map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | M. tuberculosis / ESX-1 / Type VII secretion system / EspB / Rv3881c / EsxA / CFP10 / ESAT-6 / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein secretion by the type VII secretion system / symbiont-mediated suppression of host autophagy / biological process involved in interaction with host / extracellular region / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) / Mycobacterium tuberculosis H37Rv (bacteria) /  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.37 Å | |||||||||
 Authors Authors | Piton J / Pojer F | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol X / Year: 2020 Journal: J Struct Biol X / Year: 2020Title: High resolution CryoEM structure of the ring-shaped virulence factor EspB from . Authors: Jérémie Piton / Florence Pojer / Soichi Wakatsuki / Cornelius Gati / Stewart T Cole /   Abstract: The EspB protein of is a 60 kDa virulence factor, implicated in conjugation and exported by the ESX-1 system of which it may also be a component. Previous attempts to obtain high-resolution maps of ...The EspB protein of is a 60 kDa virulence factor, implicated in conjugation and exported by the ESX-1 system of which it may also be a component. Previous attempts to obtain high-resolution maps of EspB by cryo-electron microscopic examination of single particles have been thwarted by severe orientation bias of the particles. This was overcome by using detergent as a surfactant thereby allowing reconstruction of the EspB structure at 3.37 Å resolution. The final structure revealed the N-terminal domain of EspB to be organized as a cylindrical heptamer with dimensions of 90 Å x 90 Å and a central channel of 45 Å diameter whereas the C-terminal domain was unstructured. New atomic insight was obtained into the helical packing required for protomer interactions and the overall electrostatic potential. The external surface is electronegatively charged while the channel is lined with electropositive patches. EspB thus has many features of a pore-like transport protein that might allow the passage of an ESX-1 substrate such as the 35 Å diameter EsxA-EsxB heterodimer or B-form DNA consistent with its proposed role in DNA uptake. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10658.map.gz emd_10658.map.gz | 230.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10658-v30.xml emd-10658-v30.xml emd-10658.xml emd-10658.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10658.png emd_10658.png | 58 KB | ||
| Masks |  emd_10658_msk_1.map emd_10658_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10658.cif.gz emd-10658.cif.gz | 6.3 KB | ||
| Others |  emd_10658_half_map_1.map.gz emd_10658_half_map_1.map.gz emd_10658_half_map_2.map.gz emd_10658_half_map_2.map.gz | 226.5 MB 226.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10658 http://ftp.pdbj.org/pub/emdb/structures/EMD-10658 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10658 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10658 | HTTPS FTP |
-Validation report
| Summary document |  emd_10658_validation.pdf.gz emd_10658_validation.pdf.gz | 1017.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10658_full_validation.pdf.gz emd_10658_full_validation.pdf.gz | 1017 KB | Display | |
| Data in XML |  emd_10658_validation.xml.gz emd_10658_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_10658_validation.cif.gz emd_10658_validation.cif.gz | 19.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10658 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10658 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10658 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10658 | HTTPS FTP |
-Related structure data
| Related structure data |  6xzcMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10658.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10658.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharped map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10658_msk_1.map emd_10658_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
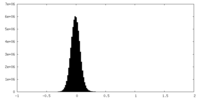
| Density Histograms |
-Half map: Map A
| File | emd_10658_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
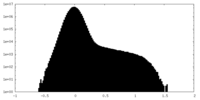
| Density Histograms |
-Half map: Map B
| File | emd_10658_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : EspB Heptameric complex
| Entire | Name: EspB Heptameric complex |
|---|---|
| Components |
|
-Supramolecule #1: EspB Heptameric complex
| Supramolecule | Name: EspB Heptameric complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 329 KDa |
-Macromolecule #1: ESX-1 secretion-associated protein EspB
| Macromolecule | Name: ESX-1 secretion-associated protein EspB / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria)Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 47.637527 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTQSQTVTVD QQEILNRANE VEAPMADPPT DVPITPCELT AAKNAAQQLV LSADNMREYL AAGAKERQRL ATSLRNAAKA YGEVDEEAA TALDNDGEGT VQAESAGAVG GDSSAELTDT PRVATAGEPN FMDLKEAARK LETGDQGASL AHFADGWNTF N LTLQGDVK ...String: MTQSQTVTVD QQEILNRANE VEAPMADPPT DVPITPCELT AAKNAAQQLV LSADNMREYL AAGAKERQRL ATSLRNAAKA YGEVDEEAA TALDNDGEGT VQAESAGAVG GDSSAELTDT PRVATAGEPN FMDLKEAARK LETGDQGASL AHFADGWNTF N LTLQGDVK RFRGFDNWEG DAATACEASL DQQRQWILHM AKLSAAMAKQ AQYVAQLHVW ARREHPTYED IVGLERLYAE NP SARDQIL PVYAEYQQRS EKVLTEYNNK AALEPVNPPK PPPAIKIDPP PPPQEQGLIP GFLMPPSDGS GVTPGTGMPA APM VPPTGS PGGGLPADTA AQLTSAGREA AALSGDVAVK AASLGGGGGG GVPSAPLGSA IGGAESVRPA GAGDIAGLGQ GRAG GGAAL GGGGMGMPMG AAHQGQGGAK SKGSQQEDEA LYTEDRAWTE AVIGNRRRQD SKESK UniProtKB: ESX-1 secretion-associated protein EspB |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 / Component:
| ||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. | ||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV Details: 0.05% fluorinated octylmaltoside was add into protein solution and immediately applied to a freshly glow-discharged holey carbon grid. ). Excess liquid was blotted for 2.3 s using an FEI ...Details: 0.05% fluorinated octylmaltoside was add into protein solution and immediately applied to a freshly glow-discharged holey carbon grid. ). Excess liquid was blotted for 2.3 s using an FEI Vitrobot Mark IV and the sample was plunge frozen in liquid ethane.. | ||||||
| Details | This sample was monodiperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 2600 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)