+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10517 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human MUC2 AAs 21-1397 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Glycoprotein / Extracellular / Polymer / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationinner mucus layer / outer mucus layer / host-mediated modulation of intestinal microbiota composition / Defective GALNT3 causes HFTC / Defective C1GALT1C1 causes TNPS / Defective GALNT12 causes CRCS1 / Termination of O-glycan biosynthesis / O-linked glycosylation of mucins / maintenance of gastrointestinal epithelium / mucus secretion ...inner mucus layer / outer mucus layer / host-mediated modulation of intestinal microbiota composition / Defective GALNT3 causes HFTC / Defective C1GALT1C1 causes TNPS / Defective GALNT12 causes CRCS1 / Termination of O-glycan biosynthesis / O-linked glycosylation of mucins / maintenance of gastrointestinal epithelium / mucus secretion / detoxification of copper ion / cupric ion binding / Dectin-2 family / cuprous ion binding / Golgi lumen / : / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
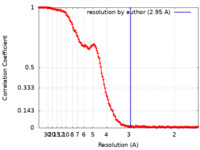
| Method | single particle reconstruction / cryo EM / Resolution: 2.95 Å | |||||||||
 Authors Authors | Javitt G / Khmelnitsky L | |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: Assembly Mechanism of Mucin and von Willebrand Factor Polymers. Authors: Gabriel Javitt / Lev Khmelnitsky / Lis Albert / Lavi Shlomo Bigman / Nadav Elad / David Morgenstern / Tal Ilani / Yaakov Levy / Ron Diskin / Deborah Fass /  Abstract: The respiratory and intestinal tracts are exposed to physical and biological hazards accompanying the intake of air and food. Likewise, the vasculature is threatened by inflammation and trauma. Mucin ...The respiratory and intestinal tracts are exposed to physical and biological hazards accompanying the intake of air and food. Likewise, the vasculature is threatened by inflammation and trauma. Mucin glycoproteins and the related von Willebrand factor guard the vulnerable cell layers in these diverse systems. Colon mucins additionally house and feed the gut microbiome. Here, we present an integrated structural analysis of the intestinal mucin MUC2. Our findings reveal the shared mechanism by which complex macromolecules responsible for blood clotting, mucociliary clearance, and the intestinal mucosal barrier form protective polymers and hydrogels. Specifically, cryo-electron microscopy and crystal structures show how disulfide-rich bridges and pH-tunable interfaces control successive assembly steps in the endoplasmic reticulum and Golgi apparatus. Remarkably, a densely O-glycosylated mucin domain performs an organizational role in MUC2. The mucin assembly mechanism and its adaptation for hemostasis provide the foundation for rational manipulation of barrier function and coagulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10517.map.gz emd_10517.map.gz | 9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10517-v30.xml emd-10517-v30.xml emd-10517.xml emd-10517.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10517_fsc.xml emd_10517_fsc.xml | 18.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10517.png emd_10517.png | 105.5 KB | ||
| Filedesc metadata |  emd-10517.cif.gz emd-10517.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10517 http://ftp.pdbj.org/pub/emdb/structures/EMD-10517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10517 | HTTPS FTP |
-Related structure data
| Related structure data |  6tm2MC  7a5oMC  6tm6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10517.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10517.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bead complex of MUC2 D1D2D3CysD1
| Entire | Name: Bead complex of MUC2 D1D2D3CysD1 |
|---|---|
| Components |
|
-Supramolecule #1: Bead complex of MUC2 D1D2D3CysD1
| Supramolecule | Name: Bead complex of MUC2 D1D2D3CysD1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Mucin-2
| Macromolecule | Name: Mucin-2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 80.356383 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SELQTEGRTR YHGRNVCSTW GNFHYKTFDG DVFRFPGLCD YNFASDCRGS YKEFAVHLKR GPGQAEAPAG VESILLTIKD DTIYLTRHL AVLNGAVVST PHYSPGLLIE KSDAYTKVYS RAGLTLMWNR EDALMLELDT KFRNHTCGLC GDYNGLQSYS E FLSDGVLF ...String: SELQTEGRTR YHGRNVCSTW GNFHYKTFDG DVFRFPGLCD YNFASDCRGS YKEFAVHLKR GPGQAEAPAG VESILLTIKD DTIYLTRHL AVLNGAVVST PHYSPGLLIE KSDAYTKVYS RAGLTLMWNR EDALMLELDT KFRNHTCGLC GDYNGLQSYS E FLSDGVLF SPLEFGNMQK INQPDVVCED PEEEVAPASC SEHRAECERL LTAEAFADCQ DLVPLEPYLR ACQQDRCRCP GG DTCVCST VAEFSRQCSH AGGRPGNWRT ATLCPKTCPG NLVYLESGSP CMDTCSHLEV SSLCEEHRMD GCFCPEGTVY DDI GDSGCV PVSQCHCRLH GHLYTPGQEI TNDCEQCVCN AGRWVCKDLP CPGTCALEGG SHITTFDGKT YTFHGDCYYV LAKG DHNDS YALLGELAPC GSTDKQTCLK TVVLLADKKK NAVVFKSDGS VLLNQLQVNL PHVTASFSVF RPSSYHIMVS MAIGV RLQV QLAPVMQLFV TLDQASQGQV QGLCGNFNGL EGDDFKTASG LVEATGAGFA NTWKAQSTCH DKLDWLDDPC SLNIES ANY AEHWCSLLKK TETPFGRCHS AVDPAEYYKR CKYDTCNCQN NEDCLCAALS SYARACTAKG VMLWGWREHV CNKDVGS CP NSQVFLYNLT TCQQTCRSLS EADSHCLEGF APVDGCGCPD HTFLDEKGRC VPLAKCSCYH RGLYLEAGDV VVRQEERC V CRDGRLHC UniProtKB: Mucin-2 |
-Macromolecule #2: Mucin-2
| Macromolecule | Name: Mucin-2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.83016 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RQIRLIGQSC TAPKIHMDCS NLTALATSKP RALSCQTLAA GYYHTECVSG CVCPDGLMDD GRGGCVVEKE CPCVHNNDLY SSGAKIKVD CNTCTCKRGR WVCTQAVCHG TCSIYGSGHY ITFDGKYYDF DGHCSYVAVQ DYCGQNSSLG SFSIITENVP C GTTGVTCS ...String: RQIRLIGQSC TAPKIHMDCS NLTALATSKP RALSCQTLAA GYYHTECVSG CVCPDGLMDD GRGGCVVEKE CPCVHNNDLY SSGAKIKVD CNTCTCKRGR WVCTQAVCHG TCSIYGSGHY ITFDGKYYDF DGHCSYVAVQ DYCGQNSSLG SFSIITENVP C GTTGVTCS KAIKIFMGRT ELKLEDKHRV VIQRDEGHHV AYTTREVGQY LVVESSTGII VIWDKRTTVF IKLAPSYKGT VC GLCGNFD HRSNNDFTTR DHMVVSSELD FGNSWKEAPT CPDVSTNPEP CSLNPHRRSW AEKQCSILKS SVFSICHSKV DPK PFYEAC VHDSCSCDTG GDCECFCSAV ASYAQECTKE GACVFWRTPD LCPIFCDYYN PPHECEWHYE PCGNRSFETC RTIN GIHSN ISVSYLEGCY PRCPKDRPIY EEDLKKCVTA DKCGCYVEDT HYP UniProtKB: Mucin-2 |
-Macromolecule #3: Mucin-2
| Macromolecule | Name: Mucin-2 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.724266 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PGASVPTEET CKSCVCTNSS QVVCRPEEGK ILNQTQDGAF CYWEICGPNG TVEKHFNICS ITTRPSTLTT FTTITLPTTP TSFTTTTTT TTPTSSTVLS TTPKLCCLWS DWINEDHPSS GSDDGDRETF DGVCGAPEDI ECRSVKDPHL SLEQHGQKVQ C DVSVGFIC ...String: PGASVPTEET CKSCVCTNSS QVVCRPEEGK ILNQTQDGAF CYWEICGPNG TVEKHFNICS ITTRPSTLTT FTTITLPTTP TSFTTTTTT TTPTSSTVLS TTPKLCCLWS DWINEDHPSS GSDDGDRETF DGVCGAPEDI ECRSVKDPHL SLEQHGQKVQ C DVSVGFIC KNEDQFGNGP FGLCYDYKIR VNCCWPMDKC ITHHHHHH UniProtKB: Mucin-2 |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 10 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 46 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 5.7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)