+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10936 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Homodimeric structure of the rBAT complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane transporter Heteromeric amino acid transporter Human transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC3A1 causes cystinuria (CSNU) / Defective SLC7A9 causes cystinuria (CSNU) / basic amino acid transport / basic amino acid transmembrane transporter activity / L-cystine transmembrane transporter activity / L-cystine transport / aspartate transmembrane transport / L-glutamate transmembrane transport / Amino acid transport across the plasma membrane / amino acid transmembrane transporter activity ...Defective SLC3A1 causes cystinuria (CSNU) / Defective SLC7A9 causes cystinuria (CSNU) / basic amino acid transport / basic amino acid transmembrane transporter activity / L-cystine transmembrane transporter activity / L-cystine transport / aspartate transmembrane transport / L-glutamate transmembrane transport / Amino acid transport across the plasma membrane / amino acid transmembrane transporter activity / vacuolar membrane / amino acid transport / brush border membrane / gene expression / carbohydrate metabolic process / apical plasma membrane / protein heterodimerization activity / protein-containing complex binding / extracellular exosome / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
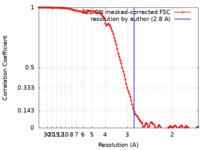
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Wu D / Safarian S | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Structural basis for amino acid exchange by a human heteromeric amino acid transporter. Authors: Di Wu / Tamara N Grund / Sonja Welsch / Deryck J Mills / Max Michel / Schara Safarian / Hartmut Michel /  Abstract: Heteromeric amino acid transporters (HATs) comprise a group of membrane proteins that belong to the solute carrier (SLC) superfamily. They are formed by two different protein components: a light ...Heteromeric amino acid transporters (HATs) comprise a group of membrane proteins that belong to the solute carrier (SLC) superfamily. They are formed by two different protein components: a light chain subunit from an SLC7 family member and a heavy chain subunit from the SLC3 family. The light chain constitutes the transport subunit whereas the heavy chain mediates trafficking to the plasma membrane and maturation of the functional complex. Mutation, malfunction, and dysregulation of HATs are associated with a wide range of pathologies or represent the direct cause of inherited and acquired disorders. Here we report the cryogenic electron microscopy structure of the neutral and basic amino acid transport complex (bAT1-rBAT) which reveals a heterotetrameric protein assembly composed of two heavy and light chain subunits, respectively. The previously uncharacterized interaction between two HAT units is mediated via dimerization of the heavy chain subunits and does not include participation of the light chain subunits. The bAT1 transporter adopts a LeuT fold and is captured in an inward-facing conformation. We identify an amino-acid-binding pocket that is formed by transmembrane helices 1, 6, and 10 and conserved among SLC7 transporters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10936.map.gz emd_10936.map.gz | 113.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10936-v30.xml emd-10936-v30.xml emd-10936.xml emd-10936.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10936_fsc.xml emd_10936_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_10936.png emd_10936.png | 132.5 KB | ||
| Filedesc metadata |  emd-10936.cif.gz emd-10936.cif.gz | 6 KB | ||
| Others |  emd_10936_half_map_1.map.gz emd_10936_half_map_1.map.gz emd_10936_half_map_2.map.gz emd_10936_half_map_2.map.gz | 96.7 MB 96.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10936 http://ftp.pdbj.org/pub/emdb/structures/EMD-10936 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10936 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10936 | HTTPS FTP |
-Related structure data
| Related structure data |  6yuzMC  6yupC  6yv1C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10936.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10936.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_10936_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10936_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homodimeric complex of rBAT
| Entire | Name: Homodimeric complex of rBAT |
|---|---|
| Components |
|
-Supramolecule #1: Homodimeric complex of rBAT
| Supramolecule | Name: Homodimeric complex of rBAT / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: 2 x rBAT |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 157.704 kDa/nm |
-Macromolecule #1: Neutral and basic amino acid transport protein rBAT
| Macromolecule | Name: Neutral and basic amino acid transport protein rBAT / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 78.937703 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAEDKSKRDS IEMSMKGCQT NNGFVHNEDI LEQTPDPGSS TDNLKHSTRG ILGSQEPDFK GVQPYAGMPK EVLFQFSGQA RYRIPREIL FWLTVASVLV LIAATIAIIA LSPKCLDWWQ EGPMYQIYPR SFKDSNKDGN GDLKGIQDKL DYITALNIKT V WITSFYKS ...String: MAEDKSKRDS IEMSMKGCQT NNGFVHNEDI LEQTPDPGSS TDNLKHSTRG ILGSQEPDFK GVQPYAGMPK EVLFQFSGQA RYRIPREIL FWLTVASVLV LIAATIAIIA LSPKCLDWWQ EGPMYQIYPR SFKDSNKDGN GDLKGIQDKL DYITALNIKT V WITSFYKS SLKDFRYGVE DFREVDPIFG TMEDFENLVA AIHDKGLKLI IDFIPNHTSD KHIWFQLSRT RTGKYTDYYI WH DCTHENG KTIPPNNWLS VYGNSSWHFD EVRNQCYFHQ FMKEQPDLNF RNPDVQEEIK EILRFWLTKG VDGFSLDAVK FLL EAKHLR DEIQVNKTQI PDTVTQYSEL YHDFTTTQVG MHDIVRSFRQ TMDQYSTEPG RYRFMGTEAY AESIDRTVMY YGLP FIQEA DFPFNNYLSM LDTVSGNSVY EVITSWMENM PEGKWPNWMI GGPDSSRLTS RLGNQYVNVM NMLLFTLPGT PITYY GEEI GMGNIVAANL NESYDINTLR SKSPMQWDNS SNAGFSEASN TWLPTNSDYH TVNVDVQKTQ PRSALKLYQD LSLLHA NEL LLNRGWFCHL RNDSHYVVYT RELDGIDRIF IVVLNFGEST LLNLHNMISG LPAKMRIRLS TNSADKGSKV DTSGIFL DK GEGLIFEHNT KNLLHRQTAF RDRCFVSNRA CYSSVLNILY TSC UniProtKB: Amino acid transporter heavy chain SLC3A1 |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 8 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Grid | Model: C-flat-1.2/1.3 4C / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)