+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10347 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
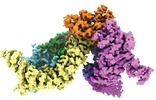
| Title | Structure of the SMG1-SMG8-SMG9 complex | |||||||||
 Map data Map data | CryoEM map of the SMG1-SMG8-SMG9 complex core | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinase / NMD / IP6 / G-fold protein / PIKK family / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of protein kinase activity / diacylglycerol-dependent serine/threonine kinase activity / chromatoid body / eye development / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / regulation of telomere maintenance / telomeric DNA binding / phosphatidylinositol phosphate biosynthetic process / mRNA export from nucleus / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) ...regulation of protein kinase activity / diacylglycerol-dependent serine/threonine kinase activity / chromatoid body / eye development / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / regulation of telomere maintenance / telomeric DNA binding / phosphatidylinositol phosphate biosynthetic process / mRNA export from nucleus / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / brain development / peptidyl-serine phosphorylation / heart development / protein autophosphorylation / in utero embryonic development / non-specific serine/threonine protein kinase / protein kinase activity / protein serine kinase activity / DNA repair / protein serine/threonine kinase activity / DNA damage response / negative regulation of apoptotic process / RNA binding / nucleoplasm / ATP binding / metal ion binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.45 Å | |||||||||
 Authors Authors | Gat Y / Schuller JM | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: InsP binding to PIKK kinases revealed by the cryo-EM structure of an SMG1-SMG8-SMG9 complex. Authors: Yair Gat / Jan Michael Schuller / Mahesh Lingaraju / Elisabeth Weyher / Fabien Bonneau / Mike Strauss / Peter J Murray / Elena Conti /   Abstract: We report the 3.45-Å resolution cryo-EM structure of human SMG1-SMG8-SMG9, a phosphatidylinositol-3-kinase (PI(3)K)-related protein kinase (PIKK) complex central to messenger RNA surveillance. ...We report the 3.45-Å resolution cryo-EM structure of human SMG1-SMG8-SMG9, a phosphatidylinositol-3-kinase (PI(3)K)-related protein kinase (PIKK) complex central to messenger RNA surveillance. Structural and MS analyses reveal the presence of inositol hexaphosphate (InsP) in the SMG1 kinase. We show that the InsP-binding site is conserved in mammalian target of rapamycin (mTOR) and potentially other PIKK members, and that it is required for optimal in vitro phosphorylation of both SMG1 and mTOR substrates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10347.map.gz emd_10347.map.gz | 3.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10347-v30.xml emd-10347-v30.xml emd-10347.xml emd-10347.xml | 25 KB 25 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10347.png emd_10347.png | 243.8 KB | ||
| Filedesc metadata |  emd-10347.cif.gz emd-10347.cif.gz | 9.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10347 http://ftp.pdbj.org/pub/emdb/structures/EMD-10347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10347 | HTTPS FTP |
-Validation report
| Summary document |  emd_10347_validation.pdf.gz emd_10347_validation.pdf.gz | 220.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10347_full_validation.pdf.gz emd_10347_full_validation.pdf.gz | 219.4 KB | Display | |
| Data in XML |  emd_10347_validation.xml.gz emd_10347_validation.xml.gz | 6.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10347 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10347 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10347 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10347 | HTTPS FTP |
-Related structure data
| Related structure data |  6sytMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10347.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10347.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of the SMG1-SMG8-SMG9 complex core | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
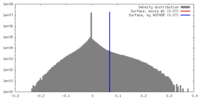
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SMG1-SMG8-SMG9 PIKK Kinase complex
| Entire | Name: SMG1-SMG8-SMG9 PIKK Kinase complex |
|---|---|
| Components |
|
-Supramolecule #1: SMG1-SMG8-SMG9 PIKK Kinase complex
| Supramolecule | Name: SMG1-SMG8-SMG9 PIKK Kinase complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: SMG1,Serine/threonine-protein kinase SMG1,SMG1,Serine/threonine-p...
| Macromolecule | Name: SMG1,Serine/threonine-protein kinase SMG1,SMG1,Serine/threonine-protein kinase SMG1,SMG1,Serine/threonine-protein kinase SMG1,SMG1,Serine/threonine-protein kinase SMG1,SMG1,Serine/threonine-protein kinase SMG1 type: protein_or_peptide / ID: 1 Details: Inactive D2335A mutant of SMG1. The automatic alignment is wrong. With the numbering here - residues 2486 to the end (from the coordinates) should be aligned with residues 3659 to the end of ...Details: Inactive D2335A mutant of SMG1. The automatic alignment is wrong. With the numbering here - residues 2486 to the end (from the coordinates) should be aligned with residues 3659 to the end of the sequence. The following residues are modeled as Alanine 147-156 162-175 191-201 207-224 248-265 267-285 290-304 311-312 1646-1657 1663-1677 1703-1717 1962-1965 1967-1978 2006-2021 2068-2083 2248-2265 Residues 1-52 are a tag. our construct has a point mutation compared to the annotated sequence - K743R,Inactive D2335A mutant of SMG1. The automatic alignment is wrong. With the numbering here - residues 2486 to the end (from the coordinates) should be aligned with residues 3659 to the end of the sequence. The following residues are modeled as Alanine 147-156 162-175 191-201 207-224 248-265 267-285 290-304 311-312 1646-1657 1663-1677 1703-1717 1962-1965 1967-1978 2006-2021 2068-2083 2248-2265 Residues 1-52 are a tag. our construct has a point mutation compared to the annotated sequence - K743R,Inactive D2335A mutant of SMG1. The automatic alignment is wrong. With the numbering here - residues 2486 to the end (from the coordinates) should be aligned with residues 3659 to the end of the sequence. The following residues are modeled as Alanine 147-156 162-175 191-201 207-224 248-265 267-285 290-304 311-312 1646-1657 1663-1677 1703-1717 1962-1965 1967-1978 2006-2021 2068-2083 2248-2265 Residues 1-52 are a tag. our construct has a point mutation compared to the annotated sequence - K743R,Inactive D2335A mutant of SMG1. The automatic alignment is wrong. With the numbering here - residues 2486 to the end (from the coordinates) should be aligned with residues 3659 to the end of the sequence. The following residues are modeled as Alanine 147-156 162-175 191-201 207-224 248-265 267-285 290-304 311-312 1646-1657 1663-1677 1703-1717 1962-1965 1967-1978 2006-2021 2068-2083 2248-2265 Residues 1-52 are a tag. our construct has a point mutation compared to the annotated sequence - K743R,Inactive D2335A mutant of SMG1. The automatic alignment is wrong. With the numbering here - residues 2486 to the end (from the coordinates) should be aligned with residues 3659 to the end of the sequence. The following residues are modeled as Alanine 147-156 162-175 191-201 207-224 248-265 267-285 290-304 311-312 1646-1657 1663-1677 1703-1717 1962-1965 1967-1978 2006-2021 2068-2083 2248-2265 Residues 1-52 are a tag. our construct has a point mutation compared to the annotated sequence - K743R,Inactive D2335A mutant of SMG1. The automatic alignment is wrong. With the numbering here - residues 2486 to the end (from the coordinates) should be aligned with residues 3659 to the end of the sequence. The following residues are modeled as Alanine 147-156 162-175 191-201 207-224 248-265 267-285 290-304 311-312 1646-1657 1663-1677 1703-1717 1962-1965 1967-1978 2006-2021 2068-2083 2248-2265 Residues 1-52 are a tag. our construct has a point mutation compared to the annotated sequence - K743R,Inactive D2335A mutant of SMG1. The automatic alignment is wrong. With the numbering here - residues 2486 to the end (from the coordinates) should be aligned with residues 3659 to the end of the sequence. The following residues are modeled as Alanine 147-156 162-175 191-201 207-224 248-265 267-285 290-304 311-312 1646-1657 1663-1677 1703-1717 1962-1965 1967-1978 2006-2021 2068-2083 2248-2265 Residues 1-52 are a tag. our construct has a point mutation compared to the annotated sequence - K743R,Inactive D2335A mutant of SMG1. The automatic alignment is wrong. With the numbering here - residues 2486 to the end (from the coordinates) should be aligned with residues 3659 to the end of the sequence. The following residues are modeled as Alanine 147-156 162-175 191-201 207-224 248-265 267-285 290-304 311-312 1646-1657 1663-1677 1703-1717 1962-1965 1967-1978 2006-2021 2068-2083 2248-2265 Residues 1-52 are a tag. our construct has a point mutation compared to the annotated sequence - K743R,Inactive D2335A mutant of SMG1. The automatic alignment is wrong. With the numbering here - residues 2486 to the end (from the coordinates) should be aligned with residues 3659 to the end of the sequence. The following residues are modeled as Alanine 147-156 162-175 191-201 207-224 248-265 267-285 290-304 311-312 1646-1657 1663-1677 1703-1717 1962-1965 1967-1978 2006-2021 2068-2083 2248-2265 Residues 1-52 are a tag. our construct has a point mutation compared to the annotated sequence - K743R,Inactive D2335A mutant of SMG1. The automatic alignment is wrong. With the numbering here - residues 2486 to the end (from the coordinates) should be aligned with residues 3659 to the end of the sequence. The following residues are modeled as Alanine 147-156 162-175 191-201 207-224 248-265 267-285 290-304 311-312 1646-1657 1663-1677 1703-1717 1962-1965 1967-1978 2006-2021 2068-2083 2248-2265 Residues 1-52 are a tag. our construct has a point mutation compared to the annotated sequence - K743R Number of copies: 1 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 401.403938 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)FSTNFR DTVDILVGWH IDHTQKPSLT QQVS GWLQS LEPFWVADLA FSTTLLGQFL EDMEAYAEDL SHVASGESVD EDVPPPSVSL PKLAALLRVF STVVRSIGER FSPIR GPPI TEAYVTDVLY RVMRCVTAAN QVFFSEAVLT AANECVGVLL GSLDPSMTIH CDMVITYGLD QLENCQTCGT DYIISV LNL LTLIVEQINT KLPSSFVEKL FIPSSKLLFL RYHKEKEVVA VAHAVYQAVL SLKNIPVLET AYKLILGEMT CALNNLL HS LQLPEACSEI KHEAFKNHVF NVDNAKFVVI FDLSALTTIG NAKNSLIGMW ALSPTVFALL SKNLMIVHSD LAVHFPAI Q YAVLYTLYSH CTRHDHFISS SLSSSSPSLF DGAVISTVTT ATKKHFSIIL NLLGILLKKD NLNQDTRKLL MTWALEAAV LMRKSETYAP LFSLPSFHKF CKGLLANTLV EDVNICLQAC SSLHALSSSL PDDLLQRCVD VCRVQLVHSG TRIRQAFGKL LKSIPLDVV LSNNNHTEIQ EISLALRSHM SKAPSNTFHP QDFSDVISFI LYGNSHRTGK DNWLERLFYS CQRLDKRDQS T IPRNLLKT DAVLWQWAIW EAAQFTVLSK LRTPLGRAQD TFQTIEGIIR SLAAHTLNPD QDVSQWTTAD NDEGHGNNQL RL VLLLQYL ENLEKLMYNA YEGCANALTS PPKVIRTFFY TNRQTCQDWL TRIRLSIMRV GLLAGQPAVT VRHGFDLLTE MKT TSLSQG NELEVTIMMV VEALCELHCP EAIQGIAVWS SSIVGKNLLW INSVAQQAEG RFEKASVEYQ EHLCAMTGVD CCIS SFDKS VLTLANAGRN SASPKHSLNG ESRKTVLSKP TDSSPEVINY LGNKACECYI SIADWAAVQE WQNAIHDLKK STSST SLNL KADFNYIKSL SSFESGKFVE CTEQLELLPG ENINLLAGGS KEKIDMKKLL PNMLSPDPRE LQKSIEVQLL RSSVCL ATA LNPIEQDQKW QSITENVVKY LKQTSRIAIG PLRLSTLTVS QSLPVLSTLQ LYCSSALENT VSNRLSTEDC LIPLFSE AL RSCKQHDVRP WMQALRYTMY QNQLLEKIKE QTVPIRSHLM ELGLTAAKFA RKRGNVSLAT RLLAQCSEVQ LGKTTTAQ D LVQHFKKLST QGQVDEKWGP ELDIEKTKLL YTAGQSTHAM EMLSSCAISF CKSVKAEYAV AKSILTLAKW IQAEWKEIS GQLKQVYRAQ HQQNFTGLST LSKNILTLIE LPSVNTMEEE YPRIESESTV HIGVGEPDFI LGQLYHLSSV QAPEVAKSWA ALASWAYRW GRKVVDNAS(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)EGVIKVWR KVVDRIFSLY KL SCSAYFT FLKLNAGQIP LDEDDPRLHL SHRVEQSTDD MIVMATLRLL RLLVKHAGEL RQYLEHGLET TPTAPWRGII PQL FSRLNH PEVYVRQSIC NLLCRVAQDS PHLILYPAIV GTISLSSESQ ASGNKFSTAI PTLLGNIQGE ELLVSECEGG SPPA SQDSN KDEPKSGLNE DQAMMQDCYS KIVDKLSSAN PTMVLQVQML VAELRRVTVL WDELWLGVLL QQHMYVLRRI QQLED EV(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)PHEK W FQDNYGDA IENALEKLK(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)YILRLEEIS PWLAAMTNTE IALPGEVSAR DTVTIHSVGG TITILPTKTK PKKLLFLGSD GKSYPYLFKG L EDLHLDER IMQFLSIVNT MFATINRQET PRFHARHYSV TPLGTRSGLI QWVDGATPLF GLYKRWQQRE AALQAQK(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)WPLHVMKA VLEELMEATP PNLLAKELWS SCTTPDE WW RVTQSYARST AVMSMVGYII GLGARHLDNV LIDMTTGEVV HIDYNVCFEK GKSLRVPEKV PFRMTQNIET ALGVTGVE G VFRLSCEQVL HIMRRGRETL LTLLEAFVYD PLVDWTAGGE AGFAGAVYGG GGQQAESKQS KREMEREITR SLFSSRVAE IKVNWFKNRD EMLVVLPKLD GSLDEYLSLQ EQLTDVEKLQ GKLLEEIEFL EGAEGVDHPS HTLQHRYSEH TQLQTQQRAV QEAIQVKLN EFEQWITHYQ AAFNNLEATQ LASLLQEIST QMDLGPPSYV PATAFLQNAG QAHLISQCEQ LEGEVGALLQ Q RRSVLRGC LEQLHHYATV ALQYPKAIFQ KHRIEQWKTW MEELICNTTV ERCQELYRKY EMQYAPQPPP TVCQFITATE MT LQRYAAD INSRLIRQVE RLKQEAVTVP VCEDQLKEIE RCIKVFLHEN GEEGSLSLAS VIISALCTLT RRNLMMEGAA SSA GEQLVD LTSRDGAWFL EELCSMSGNV TCLVQLLKQC HLVPQDLDIP NPMEASETVH LANGVYTSLQ ELNSNFRQII FPEA LRCLM KGEYTLESML HELDGLIEQT TDGVPLQTLV ESLQAYLRNA AMGLEEETHA HYIDVARLLH AQYGELIQPR NGSVD ETPK MSAGQMLLVA FDGMFAQVET AFSLLVEKLN KMEIPIAWRK IDIIREARST QVNFFDDDNH RQVLEEIFFL KRLQTI KEF FRLCGTFSKT LSGSSSLEDQ NTVNGPVQIV NVKTLFRNSC FSEDQMAKPI KAFTADFVRQ LLIGLPNQAL GLTLCSF IS ALGVDIIAQV EAKDFGAESK VSVDDLCKKA VEHNIQIGKF SQLVMNRATV LASSYDTAWK KHDLVRRLET SISSCKTS L QRVQLHIAMF QWQHEDLLIN RPQAMSVTPP PRSAILTSMK KKLHTLSQIE TSIATVQEKL AALESSIEQR LKWAGGANP ALAPVLQDFE ATIAERRNLV LKESQRASQV TFLCSNIIHF ESLRTRTAEA LNLDAALFEL IKRCQQMCSF ASQFNSSVSE LELRLLQRV DTGLEHPIGS SEWLLSAHKQ LTQDMSTQRA IQTEKEQQIE TVCETIQNLV DNIKTVLTGH NRQLGDVKHL L KAMAKDEE AALADGEDVP YENSVRQFLG EYKSWQDNIQ TVLFTLVQAM GQVRSQEHVE MLQEITPTLK ELKTQSQSIY NN LVSFASP LVTDATNECS SPTSSATYQP SFAAAVRSNT GQKTQPDVMS QNARKLIQKN LATSADTPPS TVPGTGKSVA CSP KKAVRD PKTGKAVQER NSYAVSVWKR VKAKLEGRDV DPNRRMSVAE QVDYVIKEAT NLDNLAQLYE GWTAWV UniProtKB: Serine/threonine-protein kinase SMG1, Serine/threonine-protein kinase SMG1, Serine/threonine-protein kinase SMG1, Serine/threonine-protein kinase SMG1, Serine/threonine-protein kinase SMG1 |
-Macromolecule #2: Protein SMG8
| Macromolecule | Name: Protein SMG8 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 109.82575 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGPVSLRDL LMGASAWMGS ESPGGSPTEG GGSAAGGPEP PWREDEICVV GIFGKTALRL NSEKFSLVNT VCDRQVFPLF RHQDPGDPG PGIRTEAGAV GEAGGAEDPG AAAGGSVRGS GAVAEGNRTE AGSQDYSLLQ AYYSQESKVL YLLLTSICDN S QLLRACRA ...String: MAGPVSLRDL LMGASAWMGS ESPGGSPTEG GGSAAGGPEP PWREDEICVV GIFGKTALRL NSEKFSLVNT VCDRQVFPLF RHQDPGDPG PGIRTEAGAV GEAGGAEDPG AAAGGSVRGS GAVAEGNRTE AGSQDYSLLQ AYYSQESKVL YLLLTSICDN S QLLRACRA LQSGEAGGGL SLPHAEAHEF WKHQEKLQCL SLLYLFSVCH ILLLVHPTCS FDITYDRVFR ALDGLRQKVL PL LKTAIKD CPVGKDWKLN CRPCPPRLLF LFQLNGALKV EPPRNQDPAH PDKPKKHSPK RRLQHALEDQ IYRIFRKSRV LTN QSINCL FTVPANQAFV YIVPGSQEED PVGMLLDQLR SHCTVKDPES LLVPAPLSGP RRYQVMRQHS RQQLSFHIDS SSSS SSGQL VDFTLREFLW QHVELVLSKK GFDDSVGRNP QPSHFELPTY QKWISAASKL YEVAIDGKEE DLGSPTGELT SKILS SIKV LEGFLDIDTK FSENRCQKAL PMAHSAYQSN LPHNYTMTVH KNQLAQALRV YSQHARGPAF HKYAMQLHED CYKFWS NGH QLCEERSLTD QHCVHKFHSL PKSGEKPEAD RNPPVLYHNS RARSTGACNC GRKQAPRDDP FDIKAANYDF YQLLEEK CC GKLDHINFPV FEPSTPDPAP AKNESSPAPP DSDADKLKEK EPQTQGESTS LSLALSLGQS TDSLGTYPAD PQAGGDNP E VHGQVEVKTE KRPNFVDRQA STVEYLPGML HSNCPKGLLP KFSSWSLVKL GPAKSYNFHT GLDQQGFIPG TNYLMPWDI VIRTRAEDEG DLDTNSWPAP NKAIPGKRSA VVMGRGRRRD DIARAFVGFE YEDSRGRRFM CSGPDKVMKV MGSGPKESAL KALNSDMPL YILSSSQGRG LKPHYAQLMR LFVVVPDAPL QIILMPQVQP GPPPCPVFYP EKQEITLPPD GLWVLRFPYA Y VTERGPCF PPKENVQLMS YKVLRGVLKA VTQ UniProtKB: Nonsense-mediated mRNA decay factor SMG8 |
-Macromolecule #3: Protein SMG9
| Macromolecule | Name: Protein SMG9 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.717473 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSESGHSQPG LYGIERRRRW KEPGSGGPQN LSGPGGRERD YIAPWERERR DASEETSTSV MQKTPIILSK PPAERSKQPP PPTAPAAPP APAPLEKPIV LMKPREEGKG PVAVTGASTP EGTAPPPPAA PAPPKGEKEG QRPTQPVYQI QNRGMGTAAP A AMDPVVGQ ...String: MSESGHSQPG LYGIERRRRW KEPGSGGPQN LSGPGGRERD YIAPWERERR DASEETSTSV MQKTPIILSK PPAERSKQPP PPTAPAAPP APAPLEKPIV LMKPREEGKG PVAVTGASTP EGTAPPPPAA PAPPKGEKEG QRPTQPVYQI QNRGMGTAAP A AMDPVVGQ AKLLPPERMK HSIKLVDDQM NWCDSAIEYL LDQTDVLVVG VLGLQGTGKS MVMSLLSANT PEEDQRTYVF RA QSAEMKE RGGNQTSGID FFITQERIVF LDTQPILSPS ILDHLINNDR KLPPEYNLPH TYVEMQSLQI AAFLFTVCHV VIV VQDWFT DLSLYRFLQT AEMVKPSTPS PSHESSSSSG SDEGTEYYPH LVFLQNKARR EDFCPRKLRQ MHLMIDQLMA HSHL RYKGT LSMLQCNVFP GLPPDFLDSE VNLFLVPFMD SEAESENPPR AGPGSSPLFS LLPGYRGHPS FQSLVSKLRS QVMSM ARPQ LSHTILTEKN WFHYAARIWD GVRKSSALAE YSRLLA UniProtKB: Nonsense-mediated mRNA decay factor SMG9 |
-Macromolecule #4: INOSITOL HEXAKISPHOSPHATE
| Macromolecule | Name: INOSITOL HEXAKISPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: IHP |
|---|---|
| Molecular weight | Theoretical: 660.035 Da |
| Chemical component information |  ChemComp-IHP: |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: PBS |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 52.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.45 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 214254 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)