+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10286 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cubic Dps-DNA crystal structure in vitro | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
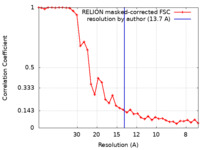
| Method | subtomogram averaging / cryo EM / Resolution: 13.7 Å | |||||||||
 Authors Authors | Chesnokov YM / Kamyshinsky RA / Vasiliev AL | |||||||||
| Funding support |  Russian Federation, 1 items Russian Federation, 1 items
| |||||||||
 Citation Citation |  Journal: Biomolecules / Year: 2019 Journal: Biomolecules / Year: 2019Title: Polymorphic Protective Dps-DNA Co-Crystals by Cryo Electron Tomography and Small Angle X-Ray Scattering. Authors: Roman Kamyshinsky / Yury Chesnokov / Liubov Dadinova / Andrey Mozhaev / Ivan Orlov / Maxim Petoukhov / Anton Orekhov / Eleonora Shtykova / Alexander Vasiliev /  Abstract: Rapid increase of intracellular synthesis of specific histone-like Dps protein that binds DNA to protect the genome against deleterious factors leads to in cellulo crystallization-one of the most ...Rapid increase of intracellular synthesis of specific histone-like Dps protein that binds DNA to protect the genome against deleterious factors leads to in cellulo crystallization-one of the most curious processes in the area of life science at the moment. However, the actual structure of the Dps-DNA co-crystals remained uncertain in the details for more than two decades. Cryo-electron tomography and small-angle X-ray scattering revealed polymorphous modifications of the co-crystals depending on the buffer parameters. Two different types of the Dps-DNA co-crystals are formed in vitro: triclinic and cubic. Three-dimensional reconstruction revealed DNA and Dps molecules in cubic co-crystals, and the unit cell parameters of cubic lattice were determined consistently by both methods. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10286.map.gz emd_10286.map.gz | 2.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10286-v30.xml emd-10286-v30.xml emd-10286.xml emd-10286.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10286_fsc.xml emd_10286_fsc.xml | 3.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_10286.png emd_10286.png | 168.1 KB | ||
| Masks |  emd_10286_msk_1.map emd_10286_msk_1.map | 2.8 MB |  Mask map Mask map | |
| Others |  emd_10286_additional.map.gz emd_10286_additional.map.gz emd_10286_additional_1.map.gz emd_10286_additional_1.map.gz emd_10286_half_map_1.map.gz emd_10286_half_map_1.map.gz emd_10286_half_map_2.map.gz emd_10286_half_map_2.map.gz | 1.9 MB 1.9 MB 1.9 MB 1.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10286 http://ftp.pdbj.org/pub/emdb/structures/EMD-10286 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10286 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10286 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10286.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10286.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10286_msk_1.map emd_10286_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
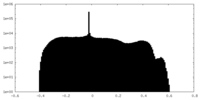
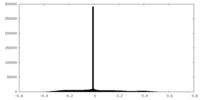
| Density Histograms |
-Additional map: Map before any post-processing
| File | emd_10286_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map before any post-processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
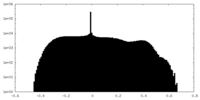
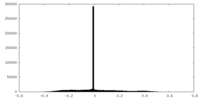
| Density Histograms |
-Additional map: Map before any post-processing
| File | emd_10286_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map before any post-processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10286_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10286_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dps-DNA co-crystal
| Entire | Name: Dps-DNA co-crystal |
|---|---|
| Components |
|
-Supramolecule #1: Dps-DNA co-crystal
| Supramolecule | Name: Dps-DNA co-crystal / type: complex / ID: 1 / Parent: 0 Details: Dps protein and DNA were mixed in the concentrations corresponding to the Dps/DNA ratio 1Dps dodecamer/60 bp of DNA. A ring vector pcDNA-hIRR-GFP 9900 bp was used as DNA sample. |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 224 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 3.1 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 / Component:
| ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: blotting for 1.5 seconds. | ||||||
| Details | weight Dps/DNA ratio = 3/1 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Cs image corrector (CEOS, Germany) |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 14.0 µm / Number grids imaged: 1 / Average exposure time: 1.0 sec. / Average electron dose: 1.02 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 37837 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 18000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)