+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10219 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The VgrG spike from the Type 6 secretion system | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Puncture / spike / tip / carrier / inhibitor / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationType VI secretion system spike protein VgrG2, C-terminal domain of unknown function DUF2345 / Putative type VI secretion system, Rhs element associated Vgr domain / Uncharacterized protein conserved in bacteria (DUF2345) / Putative type VI secretion system Rhs element Vgr / Type VI secretion system, RhsGE-associated Vgr family subset / Type VI secretion system, RhsGE-associated Vgr protein / Phage tail baseplate hub (GPD) / Gp5/Type VI secretion system Vgr protein, OB-fold domain / Type VI secretion system/phage-baseplate injector OB domain / Vgr protein, OB-fold domain superfamily Similarity search - Domain/homology | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.3 Å | |||||||||
 Authors Authors | Rapisarda C / Fronzes R | |||||||||
| Funding support |  France, France,  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2020 Journal: EMBO J / Year: 2020Title: Structural basis for loading and inhibition of a bacterial T6SS phospholipase effector by the VgrG spike. Authors: Nicolas Flaugnatti / Chiara Rapisarda / Martial Rey / Solène G Beauvois / Viet Anh Nguyen / Stéphane Canaan / Eric Durand / Julia Chamot-Rooke / Eric Cascales / Rémi Fronzes / Laure Journet /  Abstract: The bacterial type VI secretion system (T6SS) is a macromolecular machine that injects effectors into prokaryotic and eukaryotic cells. The mode of action of the T6SS is similar to contractile phages: ...The bacterial type VI secretion system (T6SS) is a macromolecular machine that injects effectors into prokaryotic and eukaryotic cells. The mode of action of the T6SS is similar to contractile phages: the contraction of a sheath structure pushes a tube topped by a spike into target cells. Effectors are loaded onto the spike or confined into the tube. In enteroaggregative Escherichia coli, the Tle1 phospholipase binds the C-terminal extension of the VgrG trimeric spike. Here, we purify the VgrG-Tle1 complex and show that a VgrG trimer binds three Tle1 monomers and inhibits their activity. Using covalent cross-linking coupled to high-resolution mass spectrometry, we provide information on the sites of contact and further identify the requirement for a Tle1 N-terminal secretion sequence in complex formation. Finally, we report the 2.6-Å-resolution cryo-electron microscopy tri-dimensional structure of the (VgrG) -(Tle1) complex revealing how the effector binds its cargo, and how VgrG inhibits Tle1 phospholipase activity. The inhibition of Tle1 phospholipase activity once bound to VgrG suggests that Tle1 dissociation from VgrG is required upon delivery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10219.map.gz emd_10219.map.gz | 153.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10219-v30.xml emd-10219-v30.xml emd-10219.xml emd-10219.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10219.png emd_10219.png | 129.9 KB | ||
| Filedesc metadata |  emd-10219.cif.gz emd-10219.cif.gz | 6.1 KB | ||
| Others |  emd_10219_additional_1.map.gz emd_10219_additional_1.map.gz emd_10219_additional_2.map.gz emd_10219_additional_2.map.gz emd_10219_half_map_1.map.gz emd_10219_half_map_1.map.gz emd_10219_half_map_2.map.gz emd_10219_half_map_2.map.gz | 408.7 KB 128.3 MB 129 MB 129 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10219 http://ftp.pdbj.org/pub/emdb/structures/EMD-10219 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10219 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10219 | HTTPS FTP |
-Related structure data
| Related structure data |  6sk0MC  6sjlC  6skiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10219.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10219.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.024 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Masked sharpened map
| File | emd_10219_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
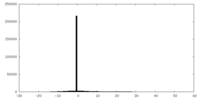
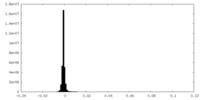
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_10219_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
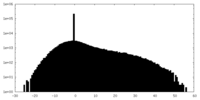
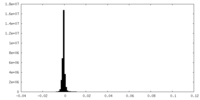
| Density Histograms |
-Half map: Half map1
| File | emd_10219_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
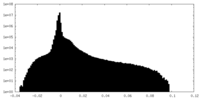
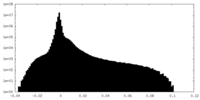
| Density Histograms |
-Half map: Half map2
| File | emd_10219_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tle1 effector bound to the VgrG spike of the type 6 secretion system
| Entire | Name: Tle1 effector bound to the VgrG spike of the type 6 secretion system |
|---|---|
| Components |
|
-Supramolecule #1: Tle1 effector bound to the VgrG spike of the type 6 secretion system
| Supramolecule | Name: Tle1 effector bound to the VgrG spike of the type 6 secretion system type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 956 KDa |
-Macromolecule #1: Putative type VI secretion protein
| Macromolecule | Name: Putative type VI secretion protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.657219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNLTDSLQNV LSGVGTLNRY RLDIPSCPSS LDVEDFSGTE GISKIYRYDI IFTSTDRYPD AAWFLRKSAT LIMGTGLLES LTEQKKVHG VITDFRRISG SEDQAQYRIT LKPFISLLDK QFRTHRFFVN KSVPEVVEQI LTEHGLKGWE YEFSLKRTYP K REQINQYQ ...String: MNLTDSLQNV LSGVGTLNRY RLDIPSCPSS LDVEDFSGTE GISKIYRYDI IFTSTDRYPD AAWFLRKSAT LIMGTGLLES LTEQKKVHG VITDFRRISG SEDQAQYRIT LKPFISLLDK QFRTHRFFVN KSVPEVVEQI LTEHGLKGWE YEFSLKRTYP K REQINQYQ ESDLRFIQRL LAEVGIFYFF TLHPDAQTEV IHFGDVQAAL IFDKTLPVNS PSGMSDSGTD SVWALNVEHR VV ESRVITN DYNHREAQNI LMSVPADMTR GEGYDTSYGE VYHYRPRHTE RGDKIDPAPE TANFWARLDH ERFLAEQTRI TGK STDASL LPAQVLTITD STPPVLPAPL QEPVLLTQLL FTGSRKSALQ VMLSAVPYSE IVCWRPPLLT RPKITGTMTA RVTS AKEGD IYAWQDASGM YRVKFDADRD DKNPGQESMP VRLAKPYSGD AYGFHFPLIQ GTEVAIAFEE GDPDRPYIAH ALHDS RHVD HVTDKNGTRN VIRTPANNKL RMEDKRGEEH IKLSTEYGGK TQLNLGHNVD ASRELRGEGA ELRTDDWISI RGGKGI FIS ADMQPQAQGK MLDMDEAIRQ LEQALSLARS MAKAATAANA TQGDISCQQR LNASLTDLTA PGMLLHAPDG IGMVSAR AL RIASGSESVG IMSGDNTDIT AGQSFTVVAE GAVSLLSRNQ GMQLLAAKGR VNIQAQSDDL SMSSQQNLDI QSSEGKVT V SANQELILAC GGAYIKLSGG NIELGCPGQI LLKSTNMQKM GPTSLDIASV EMPRGFGGGF ILTDEAGVPQ PSTPYRLTT AEGDILQGIT DENGKTAPVN TSIPSVVKVE FGKVKIHGET E UniProtKB: Putative type VI secretion protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.28 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: BACKBONE TRACE |
|---|---|
| Output model |  PDB-6sk0: |
 Movie
Movie Controller
Controller











 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)