[English] 日本語
 Yorodumi
Yorodumi- EMDB-10217: Cryo-EM structure of the RecBCD no Chi negative control complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10217 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the RecBCD no Chi negative control complex | |||||||||
 Map data Map data | EM map of the RecBCD no Chi control structure | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA repair / Homologous recombination / ATP hydrolysis / Helicase / Nuclease / translocation / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationexodeoxyribonuclease V / exodeoxyribonuclease V activity / exodeoxyribonuclease V complex / clearance of foreign intracellular DNA / DNA translocase activity / DNA 5'-3' helicase / single-stranded DNA helicase activity / recombinational repair / DNA 3'-5' helicase / 3'-5' DNA helicase activity ...exodeoxyribonuclease V / exodeoxyribonuclease V activity / exodeoxyribonuclease V complex / clearance of foreign intracellular DNA / DNA translocase activity / DNA 5'-3' helicase / single-stranded DNA helicase activity / recombinational repair / DNA 3'-5' helicase / 3'-5' DNA helicase activity / ATP-dependent activity, acting on DNA / DNA helicase activity / DNA endonuclease activity / response to radiation / helicase activity / double-strand break repair via homologous recombination / DNA recombination / 5'-3' DNA helicase activity / DNA damage response / magnesium ion binding / ATP hydrolysis activity / DNA binding / ATP binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
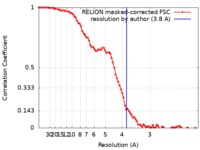
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Cheng K / Wilkinson M | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair. Authors: Kaiying Cheng / Martin Wilkinson / Yuriy Chaban / Dale B Wigley /  Abstract: The RecBCD complex plays key roles in phage DNA degradation, CRISPR array acquisition (adaptation) and host DNA repair. The switch between these roles is regulated by a DNA sequence called Chi. We ...The RecBCD complex plays key roles in phage DNA degradation, CRISPR array acquisition (adaptation) and host DNA repair. The switch between these roles is regulated by a DNA sequence called Chi. We report cryo-EM structures of the Escherichia coli RecBCD complex bound to several different DNA forks containing a Chi sequence, including one in which Chi is recognized and others in which it is not. The Chi-recognized structure shows conformational changes in regions of the protein that contact Chi and reveals a tortuous path taken by the DNA. Sequence specificity arises from interactions with both the RecC subunit and the sequence itself. These structures provide molecular details for how Chi is recognized and insights into the changes that occur in response to Chi binding that switch RecBCD from bacteriophage destruction and CRISPR spacer acquisition to constructive host DNA repair. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10217.map.gz emd_10217.map.gz | 48.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10217-v30.xml emd-10217-v30.xml emd-10217.xml emd-10217.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10217_fsc.xml emd_10217_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_10217.png emd_10217.png | 90.3 KB | ||
| Filedesc metadata |  emd-10217.cif.gz emd-10217.cif.gz | 8.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10217 http://ftp.pdbj.org/pub/emdb/structures/EMD-10217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10217 | HTTPS FTP |
-Related structure data
| Related structure data |  6sjgMC  6sjbC  6sjeC  6sjfC  6t2uC  6t2vC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10217.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10217.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map of the RecBCD no Chi control structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.048 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RecBCD bound to a forked DNA substrate that does not contain Chi
| Entire | Name: RecBCD bound to a forked DNA substrate that does not contain Chi |
|---|---|
| Components |
|
-Supramolecule #1: RecBCD bound to a forked DNA substrate that does not contain Chi
| Supramolecule | Name: RecBCD bound to a forked DNA substrate that does not contain Chi type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 355 KDa |
-Supramolecule #2: RecBCD
| Supramolecule | Name: RecBCD / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: RecBCD enzyme subunit RecB
| Macromolecule | Name: RecBCD enzyme subunit RecB / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: exodeoxyribonuclease V |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 134.123688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GMSDVAETLD PLRLPLQGER LIEASAGTGK TFTIAALYLR LLLGLGGSAA FPRPLTVEEL LVVTFTEAAT AELRGRIRSN IHELRIACL RETTDNPLYE RLLEEIDDKA QAAQWLLLAE RQMDEAAVFT IHGFCQRMLN LNAFESGMLF EQQLIEDESL L RYQACADF ...String: GMSDVAETLD PLRLPLQGER LIEASAGTGK TFTIAALYLR LLLGLGGSAA FPRPLTVEEL LVVTFTEAAT AELRGRIRSN IHELRIACL RETTDNPLYE RLLEEIDDKA QAAQWLLLAE RQMDEAAVFT IHGFCQRMLN LNAFESGMLF EQQLIEDESL L RYQACADF WRRHCYPLPR EIAQVVFETW KGPQALLRDI NRYLQGEAPV IKAPPPDDET LASRHAQIVA RIDTVKQQWR DA VGELDAL IESSGIDRRK FNRSNQAKWI DKISAWAEEE TNSYQLPESL EKFSQRFLED RTKAGGETPR HPLFEAIDQL LAE PLSIRD LVITRALAEI RETVAREKRR RGELGFDDML SRLDSALRSE SGEVLAAAIR TRFPVAMIDE FQDTDPQQYR IFRR IWHHQ PETALLLIGD PKQAIYAFRG ADIFTYMKAR SEVHAHYTLD TNWRSAPGMV NSVNKLFSQT DDAFMFREIP FIPVK SAGK NQALRFVFKG ETQPAMKMWL MEGESCGVGD YQSTMAQVCA AQIRDWLQAG QRGEALLMNG DDARPVRASD ISVLVR SRQ EAAQVRDALT LLEIPSVYLS NRDSVFETLE AQEMLWLLQA VMTPERENTL RSALATSMMG LNALDIETLN NDEHAWD VV VEEFDGYRQI WRKRGVMPML RALMSARNIA ENLLATAGGE RRLTDILHIS ELLQEAGTQL ESEHALVRWL SQHILEPD S NASSQQMRLE SDKHLVQIVT IHKSKGLEYP LVWLPFITNF RVQEQAFYHD RHSFEAVLDL NAAPESVDLA EAERLAEDL RLLYVALTRS VWHCSLGVAP LVRRRGDKKG DTDVHQSALG RLLQKGEPQD AAGLRTCIEA LCDDDIAWQT AQTGDNQPWQ VNDVSTAEL NAKTLQRLPG DNWRVTSYSG LQQRGHGIAQ DLMPRLDVDA AGVASVVEEP TLTPHQFPRG ASPGTFLHSL F EDLDFTQP VDPNWVREKL ELGGFESQWE PVLTEWITAV LQAPLNETGV SLSQLSARNK QVEMEFYLPI SEPLIASQLD TL IRQFDPL SAGCPPLEFM QVRGMLKGFI DLVFRHEGRY YLLAYKSNWL GEDSSAYTQQ AMAAAMQAHR YDLQYQLYTL ALH RYLRHR IADYDYEHHF GGVIYLFLRG VDKEHPQQGI YTTRPNAGLI ALMDEMFAGM TLEEA UniProtKB: RecBCD enzyme subunit RecB |
-Macromolecule #2: RecBCD enzyme subunit RecC
| Macromolecule | Name: RecBCD enzyme subunit RecC / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: exodeoxyribonuclease V |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 128.974102 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLRVYHSNRL DVLEALMEFI VERERLDDPF EPEMILVQST GMAQWLQMTL SQKFGIAANI DFPLPASFIW DMFVRVLPEI PKESAFNKQ SMSWKLMTLL PQLLEREDFT LLRHYLTDDS DKRKLFQLSS KAADLFDQYL VYRPDWLAQW ETGHLVEGLG E AQAWQAPL ...String: MLRVYHSNRL DVLEALMEFI VERERLDDPF EPEMILVQST GMAQWLQMTL SQKFGIAANI DFPLPASFIW DMFVRVLPEI PKESAFNKQ SMSWKLMTLL PQLLEREDFT LLRHYLTDDS DKRKLFQLSS KAADLFDQYL VYRPDWLAQW ETGHLVEGLG E AQAWQAPL WKALVEYTHQ LGQPRWHRAN LYQRFIETLE SATTCPPGLP SRVFICGISA LPPVYLQALQ ALGKHIEIHL LF TNPCRYY WGDIKDPAYL AKLLTRQRRH SFEDRELPLF RDSENAGQLF NSDGEQDVGN PLLASWGKLG RDYIYLLSDL ESS QELDAF VDVTPDNLLH NIQSDILELE NRAVAGVNIE EFSRSDNKRP LDPLDSSITF HVCHSPQREV EVLHDRLLAM LEED PTLTP RDIIVMVADI DSYSPFIQAV FGSAPADRYL PYAISDRRAR QSHPVLEAFI SLLSLPDSRF VSEDVLALLD VPVLA ARFD ITEEGLRYLR QWVNESGIRW GIDDDNVREL ELPATGQHTW RFGLTRMLLG YAMESAQGEW QSVLPYDESS GLIAEL VGH LASLLMQLNI WRRGLAQERP LEEWLPVCRD MLNAFFLPDA ETEAAMTLIE QQWQAIIAEG LGAQYGDAVP LSLLRDE LA QRLDQERISQ RFLAGPVNIC TLMPMRSIPF KVVCLLGMND GVYPRQLAPL GFDLMSQKPK RGDRSRRDDD RYLFLEAL I SAQQKLYISY IGRSIQDNSE RFPSVLVQEL IDYIGQSHYL PGDEALNCDE SEARVKAHLT CLHTRMPFDP QNYQPGERQ SYAREWLPAA SQAGKAHSEF VQPLPFTLPE TVPLETLQRF WAHPVRAFFQ MRLQVNFRTE DSEIPDTEPF ILEGLSRYQI NQQLLNALV EQDDAERLFR RFRAAGDLPY GAFGEIFWET QCQEMQQLAD RVIACRQPGQ SMEIDLACNG VQITGWLPQV Q PDGLLRWR PSLLSVAQGM QLWLEHLVYC ASGGNGESRL FLRKDGEWRF PPLAAEQALH YLSQLIEGYR EGMSAPLLVL PE SGGAWLK TCYDAQNDAM LDDDSTLQKA RTKFLQAYEG NMMVRGEGDD IWYQRLWRQL TPETMEAIVE QSQRFLLPLF RFN QS UniProtKB: RecBCD enzyme subunit RecC |
-Macromolecule #3: RecBCD enzyme subunit RecD
| Macromolecule | Name: RecBCD enzyme subunit RecD / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: exodeoxyribonuclease V |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.990367 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKLQKQLLEA VEHKQLRPLD VQFALTVAGD EHPAVTLAAA LLSHDAGEGH VCLPLSRLEN NEASHPLLAT CVSEIGELQN WEECLLASQ AVSRGDEPTP MILCGDRLYL NRMWCNERTV ARFFNEVNHA IEVDEALLAQ TLDKLFPVSD EINWQKVAAA V ALTRRISV ...String: MKLQKQLLEA VEHKQLRPLD VQFALTVAGD EHPAVTLAAA LLSHDAGEGH VCLPLSRLEN NEASHPLLAT CVSEIGELQN WEECLLASQ AVSRGDEPTP MILCGDRLYL NRMWCNERTV ARFFNEVNHA IEVDEALLAQ TLDKLFPVSD EINWQKVAAA V ALTRRISV ISGGPGTGKT TTVAKLLAAL IQMADGERCR IRLAAPTGKA AARLTESLGK ALRQLPLTDE QKKRIPEDAS TL HRLLGAQ PGSQRLRHHA GNPLHLDVLV VDEASMIDLP MMSRLIDALP DHARVIFLGD RDQLASVEAG AVLGDICAYA NAG FTAERA RQLSRLTGTH VPAGTGTEAA SLRDSLCLLQ KSYRFGSDSG IGQLAAAINR GDKTAVKTVF QQDFTDIEKR LLQS GEDYI AMLEEALAGY GRYLDLLQAR AEPDLIIQAF NEYQLLCALR EGPFGVAGLN ERIEQFMQQK RKIHRHPHSR WYEGR PVMI ARNDSALGLF NGDIGIALDR GQGTRVWFAM PDGNIKSVQP SRLPEHETTW AMTVHKSQGS EFDHAALILP SQRTPV VTR ELVYTAVTRA RRRLSLYADE RILSAAIATR TERRSGLAAL FSSRE UniProtKB: RecBCD enzyme subunit RecD |
-Macromolecule #4: Forked DNA substrate
| Macromolecule | Name: Forked DNA substrate / type: dna / ID: 4 Details: We generated a forked DNA substrate with single-stranded overhangs by annealing two synthesised oligonucleotides Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 26.049602 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DG)(DA)(DG)(DC)(DG) (DA)(DC)(DT)(DG)(DC)(DA)(DC)(DT)(DA) (DC)(DA)(DA)(DC)(DA)(DG)(DA)(DA)(DC)(DC) (DA) (DT)(DG)(DG)(DT)(DT)(DC) ...String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DG)(DA)(DG)(DC)(DG) (DA)(DC)(DT)(DG)(DC)(DA)(DC)(DT)(DA) (DC)(DA)(DA)(DC)(DA)(DG)(DA)(DA)(DC)(DC) (DA) (DT)(DG)(DG)(DT)(DT)(DC)(DT)(DG) (DT)(DT)(DG)(DT)(DA)(DG)(DT)(DG)(DC)(DA) (DG)(DT) (DC)(DG)(DC)(DT)(DC)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 1s blot before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 788 / Average exposure time: 7.0 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 47710 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)