+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10141 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of human thyroglobulin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Thyroglobulin / Thyroid / thyroid hormones / Tri-iodo-thyronine / Thyroxine / HORMONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationiodide transport / hormone biosynthetic process / thyroid hormone generation / regulation of myelination / thyroid gland development / hormone activity / signal transduction / extracellular space / extracellular region / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
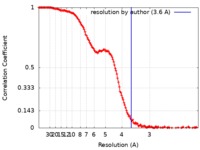
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Coscia F / Lowe J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: The structure of human thyroglobulin. Authors: Francesca Coscia / Ajda Taler-Verčič / Veronica T Chang / Ludwig Sinn / Francis J O'Reilly / Thierry Izoré / Miha Renko / Imre Berger / Juri Rappsilber / Dušan Turk / Jan Löwe /    Abstract: Thyroglobulin (TG) is the protein precursor of thyroid hormones, which are essential for growth, development and the control of metabolism in vertebrates. Hormone synthesis from TG occurs in the ...Thyroglobulin (TG) is the protein precursor of thyroid hormones, which are essential for growth, development and the control of metabolism in vertebrates. Hormone synthesis from TG occurs in the thyroid gland via the iodination and coupling of pairs of tyrosines, and is completed by TG proteolysis. Tyrosine proximity within TG is thought to enable the coupling reaction but hormonogenic tyrosines have not been clearly identified, and the lack of a three-dimensional structure of TG has prevented mechanistic understanding. Here we present the structure of full-length human thyroglobulin at a resolution of approximately 3.5 Å, determined by cryo-electron microscopy. We identified all of the hormonogenic tyrosine pairs in the structure, and verified them using site-directed mutagenesis and in vitro hormone-production assays using human TG expressed in HEK293T cells. Our analysis revealed that the proximity, flexibility and solvent exposure of the tyrosines are the key characteristics of hormonogenic sites. We transferred the reaction sites from TG to an engineered tyrosine donor-acceptor pair in the unrelated bacterial maltose-binding protein (MBP), which yielded hormone production with an efficiency comparable to that of TG. Our study provides a framework to further understand the production and regulation of thyroid hormones. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10141.map.gz emd_10141.map.gz | 12.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10141-v30.xml emd-10141-v30.xml emd-10141.xml emd-10141.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10141_fsc.xml emd_10141_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_10141.png emd_10141.png | 121.3 KB | ||
| Filedesc metadata |  emd-10141.cif.gz emd-10141.cif.gz | 8.6 KB | ||
| Others |  emd_10141_additional_1.map.gz emd_10141_additional_1.map.gz emd_10141_additional_2.map.gz emd_10141_additional_2.map.gz | 7.4 MB 14.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10141 http://ftp.pdbj.org/pub/emdb/structures/EMD-10141 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10141 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10141 | HTTPS FTP |
-Related structure data
| Related structure data |  6scjMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10666 (Title: Human thyroglobulin / Data size: 1.7 TB EMPIAR-10666 (Title: Human thyroglobulin / Data size: 1.7 TBData #1: un-aligned frames for endogenous human thyroglobulin (eTG) on graphene oxide grids (Krios 300 kV, K2 camera) [micrographs - multiframe] Data #2: un-aligned frames for recombinant human thyroglobulin produced in HEK cells (rTG) on graphene oxide grids (Krios 300 kV, K2 camera) [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10141.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10141.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.043 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_10141_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_10141_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human thyroglobulin
| Entire | Name: human thyroglobulin |
|---|---|
| Components |
|
-Supramolecule #1: human thyroglobulin
| Supramolecule | Name: human thyroglobulin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: dimeric |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 660 KDa |
-Macromolecule #1: Thyroglobulin
| Macromolecule | Name: Thyroglobulin / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 305.069844 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MALVLEIFTL LASICWVSAN IFEYQVDAQP LRPCELQRET AFLKQADYVP QCAEDGSFQT VQCQNDGRSC WCVGANGSEV LGSRQPGRP VACLSFCQLQ KQQILLSGYI NSTDTSYLPQ CQDSGDYAPV QCDVQQVQCW CVDAEGMEVY GTRQLGRPKR C PRSCEIRN ...String: MALVLEIFTL LASICWVSAN IFEYQVDAQP LRPCELQRET AFLKQADYVP QCAEDGSFQT VQCQNDGRSC WCVGANGSEV LGSRQPGRP VACLSFCQLQ KQQILLSGYI NSTDTSYLPQ CQDSGDYAPV QCDVQQVQCW CVDAEGMEVY GTRQLGRPKR C PRSCEIRN RRLLHGVGDK SPPQCSAEGE FMPVQCKFVN TTDMMIFDLV HSYNRFPDAF VTFSSFQRRF PEVSGYCHCA DS QGRELAE TGLELLLDEI YDTIFAGLDL PSTFTETTLY RILQRRFLAV QSVISGRFRC PTKCEVERFT ATSFGHPYVP SCR RNGDYQ AVQCQTEGPC WCVDAQGKEM HGTRQQGEPP SCAEGQSCAS ERQQALSRLY FGTSGYFSQH DLFSSPEKRW ASPR VARFA TSCPPTIKEL FVDSGLLRPM VEGQSQQFSV SENLLKEAIR AIFPSRGLAR LALQFTTNPK RLQQNLFGGK FLVNV GQFN LSGALGTRGT FNFSQFFQQL GLASFLNGGR QEDLAKPLSV GLDSNSSTGT PEAAKKDGTM NKPTVGSFGF EINLQE NQN ALKFLASLLE LPEFLLFLQH AISVPEDVAR DLGDVMETVL SSQTCEQTPE RLFVPSCTTE GSYEDVQCFS GECWCVN SW GKELPGSRVR GGQPRCPTDC EKQRARMQSL MGSQPAGSTL FVPACTSEGH FLPVQCFNSE CYCVDAEGQA IPGTRSAI G KPKKCPTPCQ LQSEQAFLRT VQALLSNSSM LPTLSDTYIP QCSTDGQWRQ VQCNGPPEQV FELYQRWEAQ NKGQDLTPA KLLVKIMSYR EAASGNFSLF IQSLYEAGQQ DVFPVLSQYP SLQDVPLAAL EGKRPQPREN ILLEPYLFWQ ILNGQLSQYP GSYSDFSTP LAHFDLRNCW CVDEAGQELE GMRSEPSKLP TCPGSCEEAK LRVLQFIRET EEIVSASNSS RFPLGESFLV A KGIRLRNE DLGLPPLFPP REAFAEQFLR GSDYAIRLAA QSTLSFYQRR RFSPDDSAGA SALLRSGPYM PQCDAFGSWE PV QCHAGTG HCWCVDEKGG FIPGSLTARS LQIPQCPTTC EKSRTSGLLS SWKQARSQEN PSPKDLFVPA CLETGEYARL QAS GAGTWC VDPASGEELR PGSSSSAQCP SLCNVLKSGV LSRRVSPGYV PACRAEDGGF SPVQCDQAQG SCWCVMDSGE EVPG TRVTG GQPACESPRC PLPFNASEVV GGTILCETIS GPTGSAMQQC QLLCRQGSWS VFPPGPLICS LESGRWESQL PQPRA CQRP QLWQTIQTQG HFQLQLPPGK MCSADYADLL QTFQVFILDE LTARGFCQIQ VKTFGTLVSI PVCNNSSVQV GCLTRE RLG VNVTWKSRLE DIPVASLPDL HDIERALVGK DLLGRFTDLI QSGSFQLHLD SKTFPAETIR FLQGDHFGTS PRTWFGC SE GFYQVLTSEA SQDGLGCVKC PEGSYSQDEE CIPCPVGFYQ EQAGSLACVP CPVGRTTISA GAFSQTHCVT DCQRNEAG L QCDQNGQYRA SQKDRGSGKA FCVDGEGRRL PWWETEAPLE DSQCLMMQKF EKVPESKVIF DANAPVAVRS KVPDSEFPV MQCLTDCTED EACSFFTVST TEPEISCDFY AWTSDNVACM TSDQKRDALG NSKATSFGSL RCQVKVRSHG QDSPAVYLKK GQGSTTTLQ KRFEPTGFQN MLSGLYNPIV FSASGANLTD AHLFCLLACD RDLCCDGFVL TQVQGGAIIC GLLSSPSVLL C NVKDWMDP SEAWANATCP GVTYDQESHQ VILRLGDQEF IKSLTPLEGT QDTFTNFQQV YLWKDSDMGS RPESMGCRKD TV PRPASPT EAGLTTELFS PVDLNQVIVN GNQSLSSQKH WLFKHLFSAQ QANLWCLSRC VQEHSFCQLA EITESASLYF TCT LYPEAQ VCDDIMESNA QGCRLILPQM PKALFRKKVI LEDKVKNFYT RLPFQKLMGI SIRNKVPMSE KSISNGFFEC ERRC DADPC CTGFGFLNVS QLKGGEVTCL TLNSLGIQMC SEENGGAWRI LDCGSPDIEV HTYPFGWYQK PIAQNNAPSF CPLVV LPSL TEKVSLDSWQ SLALSSVVVD PSIRHFDVAH VSTAATSNFS AVRDLCLSEC SQHEACLITT LQTQPGAVRC MFYADT QSC THSLQGQNCR LLLREEATHI YRKPGISLLS YEASVPSVPI STHGRLLGRS QAIQVGTSWK QVDQFLGVPY AAPPLAE RR FQAPEPLNWT GSWDASKPRA SCWQPGTRTS TSPGVSEDCL YLNVFIPQNV APNASVLVFF HNTMDREESE GWPAIDGS F LAAVGNLIVV TASYRVGVFG FLSSGSGEVS GNWGLLDQVA ALTWVQTHIR GFGGDPRRVS LAADRGGADV ASIHLLTAR ATNSQLFRRA VLMGGSALSP AAVISHERAQ QQAIALAKEV SCPMSSSQEV VSCLRQKPAN VLNDAQTKLL AVSGPFHYWG PVIDGHFLR EPPARALKRS LWVEVDLLIG SSQDDGLINR AKAVKQFEES RGRTSSKTAF YQALQNSLGG EDSDARVEAA A TWYYSLEH STDDYASFSR ALENATRDYF IICPIIDMAS AWAKRARGNV FMYHAPENYG HGSLELLADV QFALGLPFYP AY EGQFSLE EKSLSLKIMQ YFSHFIRSGN PNYPYEFSRK VPTFATPWPD FVPRAGGENY KEFSELLPNR QGLKKADCSF WSK YISSLK TSADGAKGGQ SAESEEEELT AGSGLREDLL SLQEPGSKTY SK UniProtKB: Thyroglobulin |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 30 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: TRIS 0.05M, 0.2M NaCl pH 8 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)