+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10097 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Faba bean necrotic stunt virus (FBNSV) | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | icosahedral T1 capsid / VIRUS | |||||||||
| Function / homology | Nanovirus coat protein / Nanovirus coat protein / T=1 icosahedral viral capsid / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Faba bean necrotic stunt virus Faba bean necrotic stunt virus | |||||||||
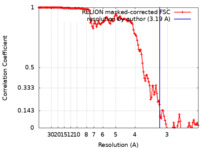
| Method | single particle reconstruction / cryo EM / Resolution: 3.19 Å | |||||||||
 Authors Authors | Trapani S / Lai Kee Him J | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2023 Journal: PLoS Pathog / Year: 2023Title: Structure-guided mutagenesis of the capsid protein indicates that a nanovirus requires assembled viral particles for systemic infection. Authors: Stefano Trapani / Eijaz Ahmed Bhat / Michel Yvon / Joséphine Lai-Kee-Him / François Hoh / Marie-Stéphanie Vernerey / Elodie Pirolles / Mélia Bonnamy / Guy Schoehn / Jean-Louis Zeddam / ...Authors: Stefano Trapani / Eijaz Ahmed Bhat / Michel Yvon / Joséphine Lai-Kee-Him / François Hoh / Marie-Stéphanie Vernerey / Elodie Pirolles / Mélia Bonnamy / Guy Schoehn / Jean-Louis Zeddam / Stéphane Blanc / Patrick Bron /  Abstract: Nanoviruses are plant multipartite viruses with a genome composed of six to eight circular single-stranded DNA segments. The distinct genome segments are encapsidated individually in icosahedral ...Nanoviruses are plant multipartite viruses with a genome composed of six to eight circular single-stranded DNA segments. The distinct genome segments are encapsidated individually in icosahedral particles that measure ≈18 nm in diameter. Recent studies on the model species Faba bean necrotic stunt virus (FBNSV) revealed that complete sets of genomic segments rarely occur in infected plant cells and that the function encoded by a given viral segment can complement the others across neighbouring cells, presumably by translocation of the gene products through unknown molecular processes. This allows the viral genome to replicate, assemble into viral particles and infect anew, even with the distinct genome segments scattered in different cells. Here, we question the form under which the FBNSV genetic material propagates long distance within the vasculature of host plants and, in particular, whether viral particle assembly is required. Using structure-guided mutagenesis based on a 3.2 Å resolution cryogenic-electron-microscopy reconstruction of the FBNSV particles, we demonstrate that specific site-directed mutations preventing capsid formation systematically suppress FBNSV long-distance movement, and thus systemic infection of host plants, despite positive detection of the mutated coat protein when the corresponding segment is agroinfiltrated into plant leaves. These results strongly suggest that the viral genome does not propagate within the plant vascular system under the form of uncoated DNA molecules or DNA:coat-protein complexes, but rather moves long distance as assembled viral particles. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10097.map.gz emd_10097.map.gz | 161.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10097-v30.xml emd-10097-v30.xml emd-10097.xml emd-10097.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10097_fsc.xml emd_10097_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_10097.png emd_10097.png | 116.5 KB | ||
| Masks |  emd_10097_msk_1.map emd_10097_msk_1.map | 172.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10097.cif.gz emd-10097.cif.gz | 5.2 KB | ||
| Others |  emd_10097_additional.map.gz emd_10097_additional.map.gz emd_10097_half_map_1.map.gz emd_10097_half_map_1.map.gz emd_10097_half_map_2.map.gz emd_10097_half_map_2.map.gz | 135.6 MB 135.9 MB 135.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10097 http://ftp.pdbj.org/pub/emdb/structures/EMD-10097 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10097 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10097 | HTTPS FTP |
-Related structure data
| Related structure data |  6s44MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10097.map.gz / Format: CCP4 / Size: 172.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10097.map.gz / Format: CCP4 / Size: 172.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.21 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10097_msk_1.map emd_10097_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened map
| File | emd_10097_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half dataset map 2
| File | emd_10097_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half dataset map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half dataset map 1
| File | emd_10097_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half dataset map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Faba bean necrotic stunt virus
| Entire | Name:  Faba bean necrotic stunt virus Faba bean necrotic stunt virus |
|---|---|
| Components |
|
-Supramolecule #1: Faba bean necrotic stunt virus
| Supramolecule | Name: Faba bean necrotic stunt virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 283824 / Sci species name: Faba bean necrotic stunt virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Name: capsid / T number (triangulation number): 3 |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Faba bean necrotic stunt virus Faba bean necrotic stunt virus |
| Molecular weight | Theoretical: 19.200422 KDa |
| Sequence | String: MVSNWNWSGK KGRRTPRRGY TRPFKSAVPT TRVVVHQSAV LKKDDVSGSE IKPEGDVARY KIRKVMLSCT LRMRPGELVN YLIVKCSSP IVNWSAAFTA PALMVKESCQ DMITIIGKGK VESNGVAGSD CTKSFNKFIR LGAGISQTQH LYVVMYTSEA V KTVLEHRV YIEV UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6s44: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)