[English] 日本語
 Yorodumi
Yorodumi- EMDB-10093: Structure of the core of the flagellar export apparatus from Vibr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10093 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the core of the flagellar export apparatus from Vibrio mimicus, the FliPQR-FlhB complex. | ||||||||||||||||||
 Map data Map data | Structure of the core of the flagellar export apparatus from Vibrio mimicus, the FliPQR-FlhB complex. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | flagella / T3SS / export apparatus / export gate / PROTEIN TRANSPORT | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationbacterial-type flagellum organization / bacterial-type flagellum basal body / bacterial-type flagellum assembly / protein secretion / protein targeting / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Vibrio mimicus CAIM 602 (bacteria) Vibrio mimicus CAIM 602 (bacteria) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||||||||
 Authors Authors | Kuhlen L / Johnson S | ||||||||||||||||||
| Funding support |  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: The substrate specificity switch FlhB assembles onto the export gate to regulate type three secretion. Authors: Lucas Kuhlen / Steven Johnson / Andreas Zeitler / Sandra Bäurle / Justin C Deme / Joseph J E Caesar / Rebecca Debo / Joseph Fisher / Samuel Wagner / Susan M Lea /   Abstract: Protein secretion through type-three secretion systems (T3SS) is critical for motility and virulence of many bacteria. Proteins are transported through an export gate containing three proteins ...Protein secretion through type-three secretion systems (T3SS) is critical for motility and virulence of many bacteria. Proteins are transported through an export gate containing three proteins (FliPQR in flagella, SctRST in virulence systems). A fourth essential T3SS protein (FlhB/SctU) functions to "switch" secretion substrate specificity once the growing hook/needle reach their determined length. Here, we present the cryo-electron microscopy structure of an export gate containing the switch protein from a Vibrio flagellar system at 3.2 Å resolution. The structure reveals that FlhB/SctU extends the helical export gate with its four predicted transmembrane helices wrapped around FliPQR/SctRST. The unusual topology of the FlhB/SctU helices creates a loop wrapped around the bottom of the closed export gate. Structure-informed mutagenesis suggests that this loop is critical in gating secretion and we propose that a series of conformational changes in the T3SS trigger opening of the gate through interactions between FlhB/SctU and FliPQR/SctRST. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10093.map.gz emd_10093.map.gz | 85 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10093-v30.xml emd-10093-v30.xml emd-10093.xml emd-10093.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10093_fsc.xml emd_10093_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_10093.png emd_10093.png | 68.8 KB | ||
| Masks |  emd_10093_msk_1.map emd_10093_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10093.cif.gz emd-10093.cif.gz | 6.5 KB | ||
| Others |  emd_10093_additional.map.gz emd_10093_additional.map.gz emd_10093_half_map_1.map.gz emd_10093_half_map_1.map.gz emd_10093_half_map_2.map.gz emd_10093_half_map_2.map.gz | 70.8 MB 71.4 MB 71.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10093 http://ftp.pdbj.org/pub/emdb/structures/EMD-10093 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10093 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10093 | HTTPS FTP |
-Related structure data
| Related structure data |  6s3lMC  6s3rC  6s3sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10093.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10093.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the core of the flagellar export apparatus from Vibrio mimicus, the FliPQR-FlhB complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10093_msk_1.map emd_10093_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
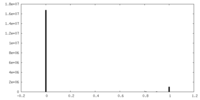
| Density Histograms |
-Additional map: refinement map
| File | emd_10093_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
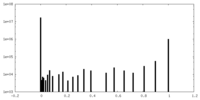
| Density Histograms |
-Half map: half1 map
| File | emd_10093_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
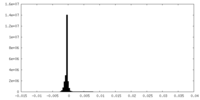
| Density Histograms |
-Half map: half2 map
| File | emd_10093_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
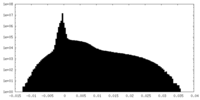
| Density Histograms |
- Sample components
Sample components
-Entire : FliPQR-FlhB
| Entire | Name: FliPQR-FlhB |
|---|---|
| Components |
|
-Supramolecule #1: FliPQR-FlhB
| Supramolecule | Name: FliPQR-FlhB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Vibrio mimicus CAIM 602 (bacteria) Vibrio mimicus CAIM 602 (bacteria) |
| Molecular weight | Theoretical: 260 KDa |
-Macromolecule #1: Flagellar biosynthetic protein FliP
| Macromolecule | Name: Flagellar biosynthetic protein FliP / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio mimicus CAIM 602 (bacteria) Vibrio mimicus CAIM 602 (bacteria) |
| Molecular weight | Theoretical: 32.431307 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKRTQRLNLT SWLTTGLLTW LLSGMLGFAS LAFAEEPLNT GIPSTAAGAS SVTVTALEKE QGNAKTIALG SSSGGSGIPA FTMTTNPDG SEDYSINLQI LALMTMLGFL PAMVILMTSF TRIVVVMSIL RQAMGLQQTP SNQVIIGIAL FLTFFIMAPV F NQINEQAV ...String: MKRTQRLNLT SWLTTGLLTW LLSGMLGFAS LAFAEEPLNT GIPSTAAGAS SVTVTALEKE QGNAKTIALG SSSGGSGIPA FTMTTNPDG SEDYSINLQI LALMTMLGFL PAMVILMTSF TRIVVVMSIL RQAMGLQQTP SNQVIIGIAL FLTFFIMAPV F NQINEQAV QPYLNEQISA RQAFDLAQEP MKAFMLKQTR IKDLETFVEM SGSQVTAPEQ VSMAVLIPAF ITSELKTAFQ IG FMLFLPF LIIDLVVASV LMAMGMMMLS PMIVSLPFKL MLFVLVDGWN LILSTLAGSF AL UniProtKB: Flagellar biosynthetic protein FliP |
-Macromolecule #2: Flagellar biosynthetic protein FliR
| Macromolecule | Name: Flagellar biosynthetic protein FliR / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio mimicus CAIM 602 (bacteria) Vibrio mimicus CAIM 602 (bacteria) |
| Molecular weight | Theoretical: 28.764324 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEYPASVVLD FIANYFWPYT RIAAMLMVMT VTGARFVPAR VRLYLGLALT FAVMPAIPAV PSDIALLSLQ GFMITFEQIV IGMAMGMVT QFLVQIFVML GQILGMQSSL GFASMVDPAN GQNTPLLGQM FMLLATLFFL SSDGHLKMIQ LVVFSFKSLP I GSGSLTTV ...String: MEYPASVVLD FIANYFWPYT RIAAMLMVMT VTGARFVPAR VRLYLGLALT FAVMPAIPAV PSDIALLSLQ GFMITFEQIV IGMAMGMVT QFLVQIFVML GQILGMQSSL GFASMVDPAN GQNTPLLGQM FMLLATLFFL SSDGHLKMIQ LVVFSFKSLP I GSGSLTTV DYRELALWLG IMFKASLAVS LSGIIALLTV NLSFGVMTRA APQLNIFSLG FSFALLVGLL LCWYILSGLY TH YEIYWQE TEEQICRLIR LNC UniProtKB: Flagellar biosynthetic protein FliR |
-Macromolecule #3: Flagellar biosynthetic protein FliQ
| Macromolecule | Name: Flagellar biosynthetic protein FliQ / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio mimicus CAIM 602 (bacteria) Vibrio mimicus CAIM 602 (bacteria) |
| Molecular weight | Theoretical: 10.333578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTPEIFVELF KESLWLVLIM VCAIIIPSLL IGLVVAIFQA ATSINEQTLS FLPRLIITLL ALMFFGHWMT QMLMDFFYSM IERLPQVLY UniProtKB: Flagellar biosynthetic protein FliQ |
-Macromolecule #4: Flagellar biosynthetic protein FlhB
| Macromolecule | Name: Flagellar biosynthetic protein FlhB / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio mimicus CAIM 602 (bacteria) Vibrio mimicus CAIM 602 (bacteria) |
| Molecular weight | Theoretical: 46.26573 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAESDGQERT EEATPRRLQQ AKEKGQVARS KELASVSVLV VGAVSLMWFG EALAQGLFTA MQRLFSLDRE EIFDIGKLFD IIGGSLVNL LLPLLMILIT LFIAALIGAA GVGGINFSAE AAMPKLSKMN PLSGFKRMFG LQSWVELLKS ILKVMLVAGV A FYLIEASQ ...String: MAESDGQERT EEATPRRLQQ AKEKGQVARS KELASVSVLV VGAVSLMWFG EALAQGLFTA MQRLFSLDRE EIFDIGKLFD IIGGSLVNL LLPLLMILIT LFIAALIGAA GVGGINFSAE AAMPKLSKMN PLSGFKRMFG LQSWVELLKS ILKVMLVAGV A FYLIEASQ KDLFQLSLDV YPQNIFHALD ILLNFVLLIS CSLLVVVAID IPFQIWQHAN QLKMTKQEVK DEYKDTEGKP EV KGRIRML QREAAQRRMM AALPQADVII TNPEHFSVAL RYKQNTDKAP VVIAKGVDHM ALKIREIARE YDIAIVPAPP LAR ALYHTT ELEQQIPDGL FVAVAQVLAF VFQLKQYRRK GGQRPKLNEE NMPIPPDMRY ENLYFQGQFG SWSHPQFEKG GGSG GGSGG GSWSHPQFEK UniProtKB: Flagellar biosynthetic protein FlhB |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: 10 seconds wait time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)