[English] 日本語
 Yorodumi
Yorodumi- EMDB-10087: Acquired functional capsid structures in metazoan totivirus-like ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10087 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Acquired functional capsid structures in metazoan totivirus-like dsRNA virus. | ||||||||||||
 Map data Map data | Omono River virus capsid | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Mosquito / Totivirus / Totiviridae / Totivirus-like virus / dsRNA / capsid / VIRUS | ||||||||||||
| Function / homology | Double-stranded RNA binding motif / Double-stranded RNA binding motif / Double stranded RNA-binding domain (dsRBD) profile. / Double-stranded RNA-binding domain / RNA binding / Capsid protein Function and homology information Function and homology information | ||||||||||||
| Biological species |  Omono River virus Omono River virus | ||||||||||||
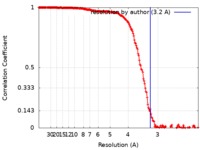
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Okamoto K / Larsson SDD | ||||||||||||
| Funding support |  Sweden, 3 items Sweden, 3 items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: Acquired Functional Capsid Structures in Metazoan Totivirus-like dsRNA Virus. Authors: Kenta Okamoto / Ricardo J Ferreira / Daniel S D Larsson / Filipe R N C Maia / Haruhiko Isawa / Kyoko Sawabe / Kazuyoshi Murata / Janos Hajdu / Kenji Iwasaki / Peter M Kasson / Naoyuki Miyazaki /    Abstract: Non-enveloped icosahedral double-stranded RNA (dsRNA) viruses possess multifunctional capsids required for their proliferation. Whereas protozoan/fungal dsRNA viruses have a relatively simple capsid ...Non-enveloped icosahedral double-stranded RNA (dsRNA) viruses possess multifunctional capsids required for their proliferation. Whereas protozoan/fungal dsRNA viruses have a relatively simple capsid structure, which suffices for the intracellular phase in their life cycle, metazoan dsRNA viruses have acquired additional structural features as an adaptation for extracellular cell-to-cell transmission in multicellular hosts. Here, we present the first atomic model of a metazoan dsRNA totivirus-like virus and the structure reveals three unique structural traits: a C-terminal interlocking arm, surface projecting loops, and an obstruction at the pore on the 5-fold symmetry axis. These traits are keys to understanding the capsid functions of metazoan dsRNA viruses, such as particle stability and formation, cell entry, and endogenous intraparticle transcription of mRNA. On the basis of molecular dynamics simulations of the obstructed pore, we propose a possible mechanism of intraparticle transcription in totivirus-like viruses, which dynamically switches between open and closed states of the pore(s). | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10087.map.gz emd_10087.map.gz | 211 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10087-v30.xml emd-10087-v30.xml emd-10087.xml emd-10087.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10087_fsc.xml emd_10087_fsc.xml | 20.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_10087.png emd_10087.png | 293.5 KB | ||
| Filedesc metadata |  emd-10087.cif.gz emd-10087.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10087 http://ftp.pdbj.org/pub/emdb/structures/EMD-10087 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10087 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10087 | HTTPS FTP |
-Validation report
| Summary document |  emd_10087_validation.pdf.gz emd_10087_validation.pdf.gz | 675.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10087_full_validation.pdf.gz emd_10087_full_validation.pdf.gz | 674.9 KB | Display | |
| Data in XML |  emd_10087_validation.xml.gz emd_10087_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_10087_validation.cif.gz emd_10087_validation.cif.gz | 22.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10087 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10087 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10087 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10087 | HTTPS FTP |
-Related structure data
| Related structure data |  6s2cMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10087.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10087.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Omono River virus capsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.12 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Omono River virus
| Entire | Name:  Omono River virus Omono River virus |
|---|---|
| Components |
|
-Supramolecule #1: Omono River virus
| Supramolecule | Name: Omono River virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Omono River virus particles from C6/36 mosquito cells NCBI-ID: 753758 / Sci species name: Omono River virus / Sci species strain: AK4 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 420.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Omono River virus Omono River virus |
| Molecular weight | Theoretical: 91.202719 KDa |
| Sequence | String: PISADFSEVE NAPSFLSLAE NTDEVLKPYT GLEIQTIITN IVGDANPNQS RIFDQDRLRG NQYSAGGLVT QNAVSAIPFT NLIPRTIRV GNILVNSANR LQITETNVSE YYSNPIIATK LSEMISDQVK NNQFSTWRRD NTSLQGFNAF DIATINTAIL P NGLSLESM ...String: PISADFSEVE NAPSFLSLAE NTDEVLKPYT GLEIQTIITN IVGDANPNQS RIFDQDRLRG NQYSAGGLVT QNAVSAIPFT NLIPRTIRV GNILVNSANR LQITETNVSE YYSNPIIATK LSEMISDQVK NNQFSTWRRD NTSLQGFNAF DIATINTAIL P NGLSLESM LLKLSLLHSI KAMNVDAASI NRSQYQVIDH NTVPTIGAPA VVGVNNSPVF GEDCGGNNPV YPFGGGTGAI AF HVTLQTV PDERKSYAIF VPPAILQATS DANEALALFA LSMSEWPHAL YTVTKQTTDL AGANAGQQVF IPTQSTIHIG GRR VLDLII PRREIAPNPT TLVAANAMCM VRPQAGPDAT AGAIPLAAGQ LFNMNFIGAP AFEEWPLTSY LYSWAGRFDI TTIR QYMGR LATMVGVKDA YWAAHELNVA LSQVAPKMTT AAGGWAAQAA NSAQQSDVCY SSLLTVTRSA ANFPLANQPA ADMRV YDTD PATWNKVALG LATAANLVPE QSMDVPFVVG DARTSFWERL QAIPMCIAWT MYYHSRGITT LAWDNAYTDN TNKWLQ KMV RNTFSTTQSV GTIIPARYGK IVCNLYKNMF HRAPAYVATS VGGKELHITH FERWLPGGTY ANVYSGAGAV VNCFSPV LI PDIWCQYFTA KLPLFAGAFP PAQGQNSTKG FNSKQGLMIH RNQNNNLVAP YLEKFADNSS YFPVGQGPEI NDMATWNG R LWMTTGNVQY LDYSGAAIVE AVPPAGELPV GKQIPLLAGE NAPIELTNAA TTCVPRYSND GRRIFTYLTT AQSVIPVQA CNRAANLARS CWLLSNVYAE PALQALGDEV EDAFDTLTNS UniProtKB: Capsid protein |
-Macromolecule #2: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Omono River virus Omono River virus |
| Molecular weight | Theoretical: 91.436969 KDa |
| Sequence | String: PISADFSEVE NAPSFLSLAE NTDEVLKPYT GLEIQTIITN IVGDANPNQS RIFDQDRLRG NQYSAGGLVT QNAVSAIPFT NLIPRTIRV GNILVNSANR LQITETNVSE YYSNPIIATK LSEMISDQVK NNQFSTWRRD NTSLQGFNAF DIATINTAIL P NGLSLESM ...String: PISADFSEVE NAPSFLSLAE NTDEVLKPYT GLEIQTIITN IVGDANPNQS RIFDQDRLRG NQYSAGGLVT QNAVSAIPFT NLIPRTIRV GNILVNSANR LQITETNVSE YYSNPIIATK LSEMISDQVK NNQFSTWRRD NTSLQGFNAF DIATINTAIL P NGLSLESM LLKLSLLHSI KAMNVDAASI NRSQYQVIDH NTVPTIGAPA VVGVNNSPVF GEDCGGNNPV YPFGGGTGAI AF HVTLQTV PDERKSYAIF VPPAILQATS DANEALALFA LSMSEWPHAL YTVTKQTTDL AGANAGQQVF IPTQSTIHIG GRR VLDLII PRREIAPNPT TLVAANAMCM VRPQAGPDAT AGAIPLAAGQ LFNMNFIGAP AFEEWPLTSY LYSWAGRFDI TTIR QYMGR LATMVGVKDA YWAAHELNVA LSQVAPKMTT AAGGWAAQAA NSAQQSDVCY SSLLTVTRSA ANFPLANQPA ADMRV YDTD PATWNKVALG LATAANLVPE QSMDVPFVVG DARTSFWERL QAIPMCIAWT MYYHSRGITT LAWDNAYTDN TNKWLQ KMV RNTFSTTQSV GTIIPARYGK IVCNLYKNMF HRAPAYVATS VGGKELHITH FERWLPGGTY ANVYSGAGAV VNCFSPV LI PDIWCQYFTA KLPLFAGAFP PAQGQNSTKG FNSKQGLMIH RNQNNNLVAP YLEKFADNSS YFPVGQGPEI NDMATWNG R LWMTTGNVQY LDYSGAAIVE AVPPAGELPV GKQIPLLAGE NAPIELTNAA TTCVPRYSND GRRIFTYLTT AQSVIPVQA CNRAANLARS CWLLSNVYAE PALQALGDEV EDAFDTLTNS SF UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.4 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Concentration: 100.0 mM / Component - Formula: C2H7NO2 / Component - Name: ammonium acetate |
| Grid | Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: COUNTING / Number real images: 4521 / Average exposure time: 1.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.225 µm / Nominal defocus min: 0.1 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-6s2c: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)