Yorodumi
Yorodumi+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1001 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Real space refinement of acto-myosin structures from sectioned muscle. | |||||||||
 Map data Map data | Acto-myosin structures from sectioned muscle | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of myosin II filament assembly / contractile muscle fiber / Striated Muscle Contraction / myosin filament / myosin complex / myosin II complex / cytoskeletal motor activator activity / myosin heavy chain binding / microfilament motor activity / tropomyosin binding ...regulation of myosin II filament assembly / contractile muscle fiber / Striated Muscle Contraction / myosin filament / myosin complex / myosin II complex / cytoskeletal motor activator activity / myosin heavy chain binding / microfilament motor activity / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / myofibril / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament / skeletal muscle thin filament assembly / actin monomer binding / skeletal muscle tissue development / skeletal muscle fiber development / stress fiber / titin binding / actin filament polymerization / muscle contraction / actin filament / filopodium / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / actin filament binding / lamellipodium / cell body / calmodulin binding / hydrolase activity / protein domain specific binding / calcium ion binding / positive regulation of gene expression / magnesium ion binding / ATP binding / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Lethocerus (insect) Lethocerus (insect) | |||||||||
| Method | electron tomography / negative staining / Resolution: 40.0 Å | |||||||||
 Authors Authors | Chen LF / Blanc E / Chapman MS / Taylor KA | |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2001 Journal: J Struct Biol / Year: 2001Title: Real space refinement of acto-myosin structures from sectioned muscle. Authors: L F Chen / E Blanc / M S Chapman / K A Taylor /  Abstract: We have adapted a real space refinement protocol originally developed for high-resolution crystallographic analysis for use in fitting atomic models of actin filaments and myosin subfragment 1 (S1) ...We have adapted a real space refinement protocol originally developed for high-resolution crystallographic analysis for use in fitting atomic models of actin filaments and myosin subfragment 1 (S1) to 3-D images of thin-sectioned, plastic-embedded whole muscle. The rationale for this effort is to obtain a refinement protocol that will optimize the fit of the model to the density obtained by electron microscopy and correct for poor geometry introduced during the manual fitting of a high-resolution atomic model into a lower resolution 3-D image. The starting atomic model consisted of a rigor acto-S1 model obtained by X-ray crystallography and helical reconstruction of electron micrographs. This model was rebuilt to fit 3-D images of rigor insect flight muscle at a resolution of 7 nm obtained by electron tomography and image averaging. Our highly constrained real space refinement resulted in modest improvements in the agreement of model and reconstruction but reduced the number of conflicting atomic contacts by 70% without loss of fit to the 3-D density. The methodology seems to be well suited to the derivation of stereochemically reasonable atomic models that are consistent with experimentally determined 3-D reconstructions computed from electron micrographs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1001.map.gz emd_1001.map.gz | 38 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1001-v30.xml emd-1001-v30.xml emd-1001.xml emd-1001.xml | 70.2 KB 70.2 KB | Display Display |  EMDB header EMDB header |
| Images |  1001.gif 1001.gif | 60.3 KB | ||
| Masks |  emd_1001_msk_1.map emd_1001_msk_1.map emd_1001_msk_10.map emd_1001_msk_10.map emd_1001_msk_11.map emd_1001_msk_11.map emd_1001_msk_12.map emd_1001_msk_12.map emd_1001_msk_13.map emd_1001_msk_13.map emd_1001_msk_14.map emd_1001_msk_14.map emd_1001_msk_15.map emd_1001_msk_15.map emd_1001_msk_16.map emd_1001_msk_16.map emd_1001_msk_17.map emd_1001_msk_17.map emd_1001_msk_18.map emd_1001_msk_18.map emd_1001_msk_19.map emd_1001_msk_19.map emd_1001_msk_2.map emd_1001_msk_2.map emd_1001_msk_20.map emd_1001_msk_20.map emd_1001_msk_21.map emd_1001_msk_21.map emd_1001_msk_22.map emd_1001_msk_22.map emd_1001_msk_23.map emd_1001_msk_23.map emd_1001_msk_24.map emd_1001_msk_24.map emd_1001_msk_25.map emd_1001_msk_25.map emd_1001_msk_26.map emd_1001_msk_26.map emd_1001_msk_3.map emd_1001_msk_3.map emd_1001_msk_4.map emd_1001_msk_4.map emd_1001_msk_5.map emd_1001_msk_5.map emd_1001_msk_6.map emd_1001_msk_6.map emd_1001_msk_7.map emd_1001_msk_7.map emd_1001_msk_8.map emd_1001_msk_8.map emd_1001_msk_9.map emd_1001_msk_9.map | 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 34.8 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB 136 KB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1001 http://ftp.pdbj.org/pub/emdb/structures/EMD-1001 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1001 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1001 | HTTPS FTP |
-Related structure data
| Related structure data |  1m8qMK  1mvwMK  1o18MK  1o19MK  1o1aMK  1o1bMK  1o1cMK  1o1dMK  1o1eMK  1o1fMK  1o1gMK M: atomic model generated by this map K: fitted to "mask" map*YM |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1001.map.gz / Format: CCP4 / Size: 40.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1001.map.gz / Format: CCP4 / Size: 40.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Acto-myosin structures from sectioned muscle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 15.4667 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
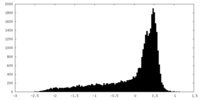
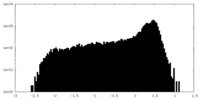
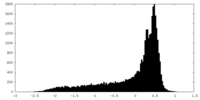
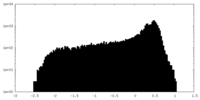
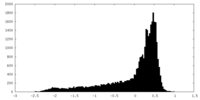
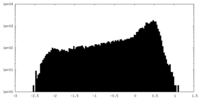
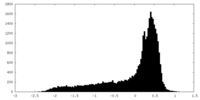
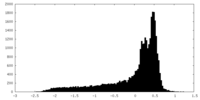
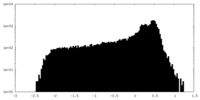
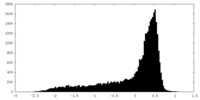
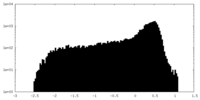
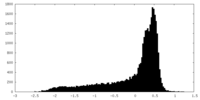
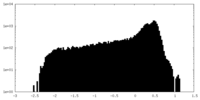
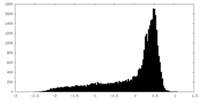
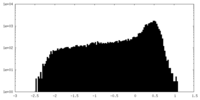
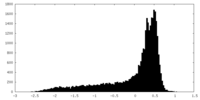
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Segmentation: #2
+Segmentation: #11
+Segmentation: #12
+Segmentation: #13
+Segmentation: #14
+Segmentation: #15
+Segmentation: #16
+Segmentation: #17
+Segmentation: #18
+Segmentation: #19
+Segmentation: #20
+Segmentation: #3
+Segmentation: #21
+Segmentation: #22
+Segmentation: #23
+Segmentation: #24
+Segmentation: #25
+Segmentation: #26
+Segmentation: #1
+Segmentation: #4
+Segmentation: #5
+Segmentation: #6
+Segmentation: #7
+Segmentation: #8
+Segmentation: #9
+Segmentation: #10
- Sample components
Sample components
-Entire : Rigor insect flight muscle from Lethocerus maximus
| Entire | Name: Rigor insect flight muscle from Lethocerus maximus |
|---|---|
| Components |
|
-Supramolecule #1000: Rigor insect flight muscle from Lethocerus maximus
| Supramolecule | Name: Rigor insect flight muscle from Lethocerus maximus / type: sample / ID: 1000 Details: The sample is a single filament layer cut from myofibrils by ultramicrotomy. The section thickness is ~25 nm. Specimen has been chemically fixed, dehydrated, embedded in Araldite and ...Details: The sample is a single filament layer cut from myofibrils by ultramicrotomy. The section thickness is ~25 nm. Specimen has been chemically fixed, dehydrated, embedded in Araldite and sectioned with a diamond knife. Oligomeric state: Thick myosin containing filament and thin actin-containing filaments. Number unique components: 2 |
|---|
-Supramolecule #1: thick, myosin-containing filament
| Supramolecule | Name: thick, myosin-containing filament / type: organelle_or_cellular_component / ID: 1 / Name.synonym: myosin / Details: Filaments contain more than just myosin / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Lethocerus (insect) / synonym: water bug, Lethocerus maximus / Tissue: Indirect flight muscles / Cell: muscle fibers / Organelle: myofibrils / Location in cell: myofibrils Lethocerus (insect) / synonym: water bug, Lethocerus maximus / Tissue: Indirect flight muscles / Cell: muscle fibers / Organelle: myofibrils / Location in cell: myofibrils |
-Supramolecule #2: thin, actin-containing filament
| Supramolecule | Name: thin, actin-containing filament / type: organelle_or_cellular_component / ID: 2 / Name.synonym: actin / Details: filaments contain more than just actin Oligomeric state: polar, 2-stranded helical filament, helical symmetry is 28/13 Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Lethocerus (insect) / synonym: water bug, Lethocerus maximus / Tissue: Indirect flight muscles / Cell: Muscle fibers / Organelle: myofibrils / Location in cell: myofibrils Lethocerus (insect) / synonym: water bug, Lethocerus maximus / Tissue: Indirect flight muscles / Cell: Muscle fibers / Organelle: myofibrils / Location in cell: myofibrils |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | electron tomography |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: NONE |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS EM400 |
|---|---|
| Temperature | Min: 25 K / Max: 25 K / Average: 25 K |
| Image recording | Digitization - Scanner: PERKIN ELMER / Digitization - Sampling interval: 25 µm / Number real images: 24 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 17000 |
| Sample stage | Specimen holder: Philips rotation-tilt holder / Specimen holder model: PHILIPS ROTATION HOLDER / Tilt series - Axis1 - Min angle: 4.3 ° / Tilt series - Axis1 - Max angle: 59.4 ° / Tilt series - Axis1 - Angle increment: 10 ° |
- Image processing
Image processing
| Details | Double axis tilt series collected on film. Digitized on PDS 1010M densitometer. Other relevent references K. A. Taylor, M. C. Reedy, L. Cordova and M. K. Reedy. 3-D structure of insect flight muscle in rigor from tilted thin sections. Nature 310, 28 5-291 (1984). Hanspeter Winkler and Kenneth A. Taylor. Multivariate statistical analysis of three-dimensional cross-bridge motifs in insect flight muscle. Ultramicroscopy 77, 141-152 (1999). Chen, Li Fan, Winkler, Hanspeter, Reedy, Michael K., Ree dy, Mary C. and Taylor, Kenneth A.. Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle. J. Struct. Biol. 138(2), 92-104 (2002) |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 40.0 Å / Resolution method: OTHER Details: The reconstruction was done by aligning the members of the tilt series using crosscorrelation functions, such as phase only and the VanHeel Mutual Correlation Function. The sampling in each ...Details: The reconstruction was done by aligning the members of the tilt series using crosscorrelation functions, such as phase only and the VanHeel Mutual Correlation Function. The sampling in each micrographof a tilted specimen was match by interpolation to th e lower angle members of the tilt series. This matching of areas and sampling was done by a 4 parameter grid search. Once aligned and scaled, the data were combined in Fourier space using a Whittaker-Shannon interpolation scheme. The map was calculated using a reverse Fourier transformation. The software used for the reconstruction was in-house and adapted from in some cases from MRC software. Number images used: 30 |
| CTF correction | Details: none |
-Atomic model buiding 1
| Software | Name: RSref |
|---|---|
| Details | Protocol: real space rigid body. Domains were fit manually into the density using O. The entire light chain domain consisting of heavy chain residues from G710 to K843 and the two light chains were repositioned using G710 of the myosin heavy chain using O.Real space refinement done using 7 rigid bodies and 4 hinge points. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT Target criteria: poor atom-atom contacts and cross correlation coefficient |
| Output model |  PDB-1m8q:  PDB-1mvw:  PDB-1o18:  PDB-1o19:  PDB-1o1a:  PDB-1o1b:  PDB-1o1c:  PDB-1o1d:  PDB-1o1e:  PDB-1o1f:  PDB-1o1g: |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)