[English] 日本語
 Yorodumi
Yorodumi- EMDB-0992: Cryo-EM structure of the multiple peptide resistance factor (MprF... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0992 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the multiple peptide resistance factor (MprF) loaded with one lysyl-phosphatidylglycerol molecule | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | bacteria membrane protein / MEMBRANE PROTEIN | ||||||||||||
| Function / homology | : / Phosphatidylglycerol lysyltransferase, C-terminal / Phosphatidylglycerol lysyltransferase, C-terminal / aminoacyltransferase activity / phospholipid homeostasis / Acyl-CoA N-acyltransferase / plasma membrane / Bifunctional lysylphosphatidylglycerol flippase/synthetase MprF / :  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Rhizobium tropici (bacteria) / Rhizobium tropici (bacteria) /  Rhizobium tropici CIAT 899 (bacteria) Rhizobium tropici CIAT 899 (bacteria) | ||||||||||||
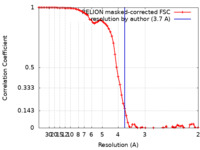
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Song DF / Jiao HZ | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Phospholipid translocation captured in a bifunctional membrane protein MprF. Authors: Danfeng Song / Haizhan Jiao / Zhenfeng Liu /  Abstract: As a large family of membrane proteins crucial for bacterial physiology and virulence, the Multiple Peptide Resistance Factors (MprFs) utilize two separate domains to synthesize and translocate ...As a large family of membrane proteins crucial for bacterial physiology and virulence, the Multiple Peptide Resistance Factors (MprFs) utilize two separate domains to synthesize and translocate aminoacyl phospholipids to the outer leaflets of bacterial membranes. The function of MprFs enables Staphylococcus aureus and other pathogenic bacteria to acquire resistance to daptomycin and cationic antimicrobial peptides. Here we present cryo-electron microscopy structures of MprF homodimer from Rhizobium tropici (RtMprF) at two different states in complex with lysyl-phosphatidylglycerol (LysPG). RtMprF contains a membrane-embedded lipid-flippase domain with two deep cavities opening toward the inner and outer leaflets of the membrane respectively. Intriguingly, a hook-shaped LysPG molecule is trapped inside the inner cavity with its head group bent toward the outer cavity which hosts a second phospholipid-binding site. Moreover, RtMprF exhibits multiple conformational states with the synthase domain adopting distinct positions relative to the flippase domain. Our results provide a detailed framework for understanding the mechanisms of MprF-mediated modification and translocation of phospholipids. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0992.map.gz emd_0992.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0992-v30.xml emd-0992-v30.xml emd-0992.xml emd-0992.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0992_fsc.xml emd_0992_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0992.png emd_0992.png | 23.4 KB | ||
| Filedesc metadata |  emd-0992.cif.gz emd-0992.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0992 http://ftp.pdbj.org/pub/emdb/structures/EMD-0992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0992 | HTTPS FTP |
-Validation report
| Summary document |  emd_0992_validation.pdf.gz emd_0992_validation.pdf.gz | 427.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0992_full_validation.pdf.gz emd_0992_full_validation.pdf.gz | 427.1 KB | Display | |
| Data in XML |  emd_0992_validation.xml.gz emd_0992_validation.xml.gz | 8.2 KB | Display | |
| Data in CIF |  emd_0992_validation.cif.gz emd_0992_validation.cif.gz | 10.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0992 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0992 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0992 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0992 | HTTPS FTP |
-Related structure data
| Related structure data |  6lvfMC  6lv0C  7duwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0992.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0992.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : bacterial membrane protein
| Entire | Name: bacterial membrane protein |
|---|---|
| Components |
|
-Supramolecule #1: bacterial membrane protein
| Supramolecule | Name: bacterial membrane protein / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: A recombinant protein from Rhizobium tropici expressed in Escherichia coli cells |
|---|---|
| Source (natural) | Organism:  Rhizobium tropici (bacteria) Rhizobium tropici (bacteria) |
-Macromolecule #1: Low pH-inducible protein LpiA
| Macromolecule | Name: Low pH-inducible protein LpiA / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhizobium tropici CIAT 899 (bacteria) Rhizobium tropici CIAT 899 (bacteria) |
| Molecular weight | Theoretical: 96.094672 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSPIDLESA DEELEERGGI VGLLNRYRAL IVAVLTVAVF CIAAYAIYDL TTEVRYDDVV HALTTTKISS VLLALLFTGL SFASLIFYD QNALEYIGKR LPFPHVALTS FSAYAVGNTA GFGALSAGAI RYRAYTRLGL SPDDITRVIA FVTLAFGLGL A SVGAMALL ...String: MSSPIDLESA DEELEERGGI VGLLNRYRAL IVAVLTVAVF CIAAYAIYDL TTEVRYDDVV HALTTTKISS VLLALLFTGL SFASLIFYD QNALEYIGKR LPFPHVALTS FSAYAVGNTA GFGALSAGAI RYRAYTRLGL SPDDITRVIA FVTLAFGLGL A SVGAMALL VIADEIGPLI SVDGLWLRLI AIAILAALAF VVYAGRNGRE VRIGPVAVRL PDSRTWSRQF LVTAFDIAAS AS VLYVLLP ETSIGWPGFF AIYAIAVGLG VLSHVPAGFG VFETIIIAWL GSSVNEDAVL SSLVLYRVIY NVIPLVIAIA AIS VAELRR FVDHPVASSM RRIGARLMPQ LLSAFALLLG MMLVFSSVTP TPDHNLIVLS DYLPLSLVES AHFLSSLLGL AIIV AARGL SQRLDGAWWV STFSALFALF FSLLKAIAIV EAGLLAFFVF SLVVSRRLFK RPASLLNQTL TAGWLTAIAV VCIGA IVVL FFVYRDVGYS NELWWQFEFA DEAPRGLRAA LGISIVSSAI AIFSLLRPAT KRPEPVSDDA VARAVEIVRK QGVADA NLV RMGDKSIMFS EKGDAFIMYG KQGRSWIALF DPVGPRQALP DLIWRFVETA RAAGCRSVFY QISPALLSYC ADAGLRA FK LGELAVVNLA NFELKGGKWA NLRQTASRAV RDGLEFAVIE PQDIPDVLDQ LAHVSDTWLA DHNAKEKSFS LGAFDPDY V CSQPVGVLKK DGKIVAFANI LMTETKEEGS VDLMRFSPDA PKGSMDFLFV QILEYLKGEG FQRFNLGMAP LSGMSRRES APVWDRVGGT VFEHGERFYN FKGLRAFKSK FHPEWQPRYL AVSGGVSPMI ALMDATFLIG GGLKGVVRKK LAAALEHHHH HH UniProtKB: UNIPROTKB: L0LM43 |
-Macromolecule #2: [(2~{R})-3-[[(2~{S})-3-[(2~{S})-2,6-bis(azanyl)hexanoyl]oxy-2-oxi...
| Macromolecule | Name: [(2~{R})-3-[[(2~{S})-3-[(2~{S})-2,6-bis(azanyl)hexanoyl]oxy-2-oxidanyl-propoxy]-oxidanyl-phosphoryl]oxy-2-hexadecanoyloxy-propyl] hexadecanoate type: ligand / ID: 2 / Number of copies: 2 / Formula: EV9 |
|---|---|
| Molecular weight | Theoretical: 851.142 Da |
| Chemical component information |  ChemComp-EV9: |
-Macromolecule #3: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
| Macromolecule | Name: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE / type: ligand / ID: 3 / Number of copies: 2 / Formula: LHG |
|---|---|
| Molecular weight | Theoretical: 722.97 Da |
| Chemical component information |  ChemComp-LHG: |
-Macromolecule #4: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
| Macromolecule | Name: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 4 / Number of copies: 2 / Formula: PGT |
|---|---|
| Molecular weight | Theoretical: 751.023 Da |
| Chemical component information |  ChemComp-PGT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | The protein is reconstituted in lipid nanodiscs |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-32 / Number grids imaged: 1 / Number real images: 2921 / Average exposure time: 5.2 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6lvf: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)