+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0946 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the mouse Piezo1 isoform Piezo1.1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Piezo1.1 / Mechanogating / Mechanotransduction channel / MEMBRANE PROTEIN / isoform | |||||||||
| Function / homology |  Function and homology information Function and homology informationmechanosensitive monoatomic cation channel activity / cuticular plate / positive regulation of cell-cell adhesion mediated by integrin / positive regulation of integrin activation / detection of mechanical stimulus / mechanosensitive monoatomic ion channel activity / stereocilium / positive regulation of myotube differentiation / lamellipodium membrane / monoatomic cation transport ...mechanosensitive monoatomic cation channel activity / cuticular plate / positive regulation of cell-cell adhesion mediated by integrin / positive regulation of integrin activation / detection of mechanical stimulus / mechanosensitive monoatomic ion channel activity / stereocilium / positive regulation of myotube differentiation / lamellipodium membrane / monoatomic cation transport / monoatomic cation channel activity / endoplasmic reticulum-Golgi intermediate compartment membrane / regulation of membrane potential / endoplasmic reticulum membrane / endoplasmic reticulum / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Geng J / Liu W / Zhou H / Zhang T / Wang L / Zhang M / Shen B / Li X / Xiao B | |||||||||
 Citation Citation |  Journal: Neuron / Year: 2020 Journal: Neuron / Year: 2020Title: A Plug-and-Latch Mechanism for Gating the Mechanosensitive Piezo Channel. Authors: Jie Geng / Wenhao Liu / Heng Zhou / Tingxin Zhang / Li Wang / Mingmin Zhang / Yiran Li / Bo Shen / Xueming Li / Bailong Xiao /  Abstract: The mechanosensitive Piezo1 and Piezo2 channels convert mechanical force into cation permeation. However, their precise mechanogating and regulatory mechanisms remain elusive. Here, we report that ...The mechanosensitive Piezo1 and Piezo2 channels convert mechanical force into cation permeation. However, their precise mechanogating and regulatory mechanisms remain elusive. Here, we report that Piezo1 utilizes three lateral ion-conducting portals equipped with physical gates for cooperative gating and splicing regulation. Mutating residues lining the portal converts Piezo1 into an anion-selective channel, demonstrating the portal-based cation-permeating pathway. Intriguingly, the portal is physically blocked with a plug domain, which undergoes alternative splicing in both Piezo1 and Piezo2. The Piezo1 isoform has local openings of the portals, enlarged single-channel conductance and sensitized mechanosensitivity. Remarkably, the three plugs are strategically latched onto the central axis for coordinated gating of the three portals. Disrupting the latching induces three quantal sub-conductance states in Piezo1, but not in the isoform. Together, we propose that Piezo utilizes an elegant plug-and-latch mechanism to physically and coordinately gate the lateral portals through the spliceable plug gates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0946.map.gz emd_0946.map.gz | 8.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0946-v30.xml emd-0946-v30.xml emd-0946.xml emd-0946.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0946.png emd_0946.png | 184.2 KB | ||
| Filedesc metadata |  emd-0946.cif.gz emd-0946.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0946 http://ftp.pdbj.org/pub/emdb/structures/EMD-0946 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0946 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0946 | HTTPS FTP |
-Related structure data
| Related structure data |  6lqiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0946.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0946.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
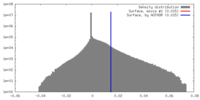
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : homo-trimer of Piezo1.1
| Entire | Name: homo-trimer of Piezo1.1 |
|---|---|
| Components |
|
-Supramolecule #1: homo-trimer of Piezo1.1
| Supramolecule | Name: homo-trimer of Piezo1.1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 880 KDa |
-Macromolecule #1: Piezo-type mechanosensitive ion channel component 1
| Macromolecule | Name: Piezo-type mechanosensitive ion channel component 1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 289.590625 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEPHVLGAGL YWLLLPCTLL AASLLRFNAL SLVYLLFLLL LPWLPGPSRH SIPGHTGRLL RALLCLSLLF LVAHLAFQIC LHTVPHLDQ FLGQNGSLWV KVSQHIGVTR LDLKDIFNTT RLVAPDLGVL LASSLCLGLC GRLTRKAGQS RRTQELQDDD D DDDDDDED ...String: MEPHVLGAGL YWLLLPCTLL AASLLRFNAL SLVYLLFLLL LPWLPGPSRH SIPGHTGRLL RALLCLSLLF LVAHLAFQIC LHTVPHLDQ FLGQNGSLWV KVSQHIGVTR LDLKDIFNTT RLVAPDLGVL LASSLCLGLC GRLTRKAGQS RRTQELQDDD D DDDDDDED IDAAPAVGLK GAPALATKRR LWLASRFRVT AHWLLMTSGR TLVIVLLALA GIAHPSAFSS IYLVVFLAIC TW WSCHFPL SPLGFNTLCV MVSCFGAGHL ICLYCYQTPF IQDMLPPGNI WARLFGLKNF VDLPNYSSPN ALVLNTKHAW PIY VSPGIL LLLYYTATSL LKLHKSCPSE LRKETPREDE EHELELDHLE PEPQARDATQ GEMPMTTEPD LDNCTVHVLT SQSP VRQRP VRPRLAELKE MSPLHGLGHL IMDQSYVCAL IAMMVWSIMY HSWLTFVLLL WACLIWTVRS RHQLAMLCSP CILLY GLTL CCLRYVWAME LPELPTTLGP VSLHQLGLEH TRYPCLDLGA MLLYLLTFWL LLRQFVKEKL LKKQKVPAAL LEVTVA DTE PTQTQTLLRS LGELVTGIYV KYWIYVCAGM FIVVSFAGRL VVYKIVYMFL FLLCLTLFQV YYTLWRKLLR VFWWLVV AY TMLVLIAVYT FQFQDFPTYW RNLTGFTDEQ LGDLGLEQFS VSELFSSILI PGFFLLACIL QLHYFHRPFM QLTDLEHV P PPGTRHPRWA HRQDAVSEAP LLEHQEEEEV FREDGQSMDG PHQATQVPEG TASKWGLVAD RLLDLAASFS AVLTRIQVF VRRLLELHVF KLVALYTVWV ALKEVSVMNL LLVVLWAFAL PYPRFRPMAS CLSTVWTCII IVCKMLYQLK IVNPHEYSSN CTEPFPNNT NLQPLEINQS LLYRGPVDPA NWFGVRKGYP NLGYIQNHLQ ILLLLVFEAV VYRRQEHYRR QHQQAPLPAQ A VCADGTRQ RLDQDLLSCL KYFINFFFYK FGLEICFLMA VNVIGQRMNF MVILHGCWLV AILTRRRREA IARLWPNYCL FL TLFLLYQ YLLCLGMPPA LCIDYPWRWS KAIPMNSALI KWLYLPDFFR APNSTNLISD FLLLLCASQQ WQVFSAERTE EWQ RMAGIN TDHLEPLRGE PNPIPNFIHC RSYLDMLKVA VFRYLFWLVL VVVFVAGATR ISIFGLGYLL ACFYLLLFGT TLLQ KDTRA QLVLWDCLIL YNVTVIISKN MLSLLSCVFV EQMQSNFCWV IQLFSLVCTV KGYYDPKEMM TRDRDCLLPV EEAGI IWDS ICFFFLLLQR RIFLSHYFLH VSADLKATAL QASRGFALYN AANLKSINFH RQIEEKSLAQ LKRQMKRIRA KQEKYR QSQ ASRGQLQSKD PQDPSQEPVI HSGDYFLFES DSEEEEEALP EDPRPAAQSA FQMAYQAWVT NAQTVLRQRR ERARQER AE QLASGGDLNP DVEPVDVPED EMAGRSHMMQ RVLSTMQFLW VLGQATVDGL TRWLRAFTKH HRTMSDVLCA ERYLLTQE L LRVGEVRRGV LDQLYVGEDE ATLSGPVETR DGPSTASSGL GAEEPLSSMT DDTSSPLSTG YNTRSGSEEI VTDAGDLQA GTSLHGSQEL LANARTRMRT ASELLLDRRL HIPELEEAER FEAQQGRTLR LLRAGYQCVA AHSELLCYFI IILNHMVTAS AASLVLPVL VFLWAMLTIP RPSKRFWMTA IVFTEVMVVT KYLFQFGFFP WNSYVVLRRY ENKPYFPPRI LGLEKTDSYI K YDLVQLMA LFFHRSQLLC YGLWDHEEDR YPKDHCRSSV KDREAKEEPE AKLESQSETG TGHPKEPVLA GTPRDHIQGK GS IRSKDVI QDPPEDLKPR HTRHISIRFR RRKETPGPKG TAVMETEHEE GEGKETTERK RPRHTQEKSK FRERMKAAGR RLQ SFCVSL AQSFYQPLQR FFHDILHTKY RAATDVYALM FLADIVDIII IIFGFWAFGK HSAATDIASS LSDDQVPQAF LFML LVQFG TMVIDRALYL RKTVLGKLAF QVVLVVAIHI WMFFILPAVT ERMFSQNAVA QLWYFVKCIY FALSAYQIRC GYPTR ILGN FLTKKYNHLN LFLFQGFRLV PFLVELRAVM DWVWTDTTLS LSNWMCVEDI YANIFIIKCS RETEKKYPQP KGQKKK KIV KYGMGGLIIL FLIAIIWFPL LFMSLIRSVV GVVNQPIDVT VTLKLGGYEP LFTMSAQQPS IVPFTPQAYE ELSQQFD PY PLAMQFISQY SPEDIVTAQI EGSSGALWRI SPPSRAQMKQ ELYNGTADIT LRFTWNFQRD LAKGGTVEYT NEKHTLEL A PNSTARRQLA QLLEGRPDQS VVIPHLFPKY IRAPNGPEAN PVKQLQPDEE EDYLGVRIQL RREQVGTGAS GEQAGTKAS DFLEWWVIEL QDCKADCNLL PMVIFSDKVS PPSLGFLAGY GIVGLYVSIV LVVGKFVRGF FSEISHSIMF EELPCVDRIL KLCQDIFLV RETRELELEE ELYAKLIFLY RSPETMIKWT RERE UniProtKB: Piezo-type mechanosensitive ion channel component 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 179654 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.0) |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6lqi: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)