[English] 日本語
 Yorodumi
Yorodumi- EMDB-0726: Structure of PSI from H. hongdechloris grown under white light co... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0726 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of PSI from H. hongdechloris grown under white light condition | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Photosystem I / ELECTRON TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationphotosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / plasma membrane-derived thylakoid membrane / chlorophyll binding / photosynthesis / endomembrane system / 4 iron, 4 sulfur cluster binding / electron transfer activity ...photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / plasma membrane-derived thylakoid membrane / chlorophyll binding / photosynthesis / endomembrane system / 4 iron, 4 sulfur cluster binding / electron transfer activity / oxidoreductase activity / magnesium ion binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Halomicronema hongdechloris C2206 (bacteria) Halomicronema hongdechloris C2206 (bacteria) | |||||||||
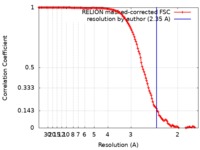
| Method | single particle reconstruction / cryo EM / Resolution: 2.35 Å | |||||||||
 Authors Authors | Kato K / Nagao R | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structural basis for the adaptation and function of chlorophyll f in photosystem I. Authors: Koji Kato / Toshiyuki Shinoda / Ryo Nagao / Seiji Akimoto / Takehiro Suzuki / Naoshi Dohmae / Min Chen / Suleyman I Allakhverdiev / Jian-Ren Shen / Fusamichi Akita / Naoyuki Miyazaki / Tatsuya Tomo /    Abstract: Chlorophylls (Chl) play pivotal roles in energy capture, transfer and charge separation in photosynthesis. Among Chls functioning in oxygenic photosynthesis, Chl f is the most red-shifted type first ...Chlorophylls (Chl) play pivotal roles in energy capture, transfer and charge separation in photosynthesis. Among Chls functioning in oxygenic photosynthesis, Chl f is the most red-shifted type first found in a cyanobacterium Halomicronema hongdechloris. The location and function of Chl f in photosystems are not clear. Here we analyzed the high-resolution structures of photosystem I (PSI) core from H. hongdechloris grown under white or far-red light by cryo-electron microscopy. The structure showed that, far-red PSI binds 83 Chl a and 7 Chl f, and Chl f are associated at the periphery of PSI but not in the electron transfer chain. The appearance of Chl f is well correlated with the expression of PSI genes induced under far-red light. These results indicate that Chl f functions to harvest the far-red light and enhance uphill energy transfer, and changes in the gene sequences are essential for the binding of Chl f. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0726.map.gz emd_0726.map.gz | 24 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0726-v30.xml emd-0726-v30.xml emd-0726.xml emd-0726.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0726_fsc.xml emd_0726_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_0726.png emd_0726.png | 97.2 KB | ||
| Filedesc metadata |  emd-0726.cif.gz emd-0726.cif.gz | 7.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0726 http://ftp.pdbj.org/pub/emdb/structures/EMD-0726 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0726 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0726 | HTTPS FTP |
-Related structure data
| Related structure data |  6kmwMC  0727C  6kmxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0726.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0726.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.87 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : white PSI
+Supramolecule #1: white PSI
+Macromolecule #1: Photosystem I P700 chlorophyll a apoprotein A1
+Macromolecule #2: Photosystem I P700 chlorophyll a apoprotein A2
+Macromolecule #3: Photosystem I iron-sulfur center
+Macromolecule #4: Photosystem I reaction center subunit II
+Macromolecule #5: Photosystem I reaction center subunit IV
+Macromolecule #6: Photosystem I reaction center subunit VIII
+Macromolecule #7: Photosystem I reaction center subunit XI
+Macromolecule #8: Photosystem I reaction center subunit XII
+Macromolecule #9: CHLOROPHYLL A ISOMER
+Macromolecule #10: CHLOROPHYLL A
+Macromolecule #11: PHYLLOQUINONE
+Macromolecule #12: IRON/SULFUR CLUSTER
+Macromolecule #13: BETA-CAROTENE
+Macromolecule #14: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #15: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #16: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #17: UNKNOWN LIGAND
+Macromolecule #18: CALCIUM ION
+Macromolecule #19: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.032 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)