+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0724 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | FimA type V pilus from P.gingivalis | ||||||||||||||||||
 Map data Map data | FimA pilus cryo-EM reconstruction | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Type V pilus / FimA / Porphyromonas gingivalis / ATCC33277 / CELL ADHESION | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpilus / cell outer membrane / cell adhesion / structural molecule activity Similarity search - Function | ||||||||||||||||||
| Biological species |  Porphyromonas gingivalis ATCC 33277 (bacteria) Porphyromonas gingivalis ATCC 33277 (bacteria) | ||||||||||||||||||
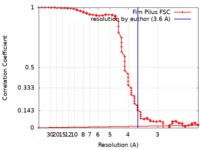
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||||||||
 Authors Authors | Shibata S / Shoji M | ||||||||||||||||||
| Funding support |  Japan, 5 items Japan, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2020 Journal: Nat Microbiol / Year: 2020Title: Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange. Authors: Satoshi Shibata / Mikio Shoji / Kodai Okada / Hideyuki Matsunami / Melissa M Matthews / Katsumi Imada / Koji Nakayama / Matthias Wolf /  Abstract: Bacterial adhesion is a general strategy for host-microbe and microbe-microbe interactions. Adhesive pili are essential for colonization, biofilm formation, virulence and pathogenesis of many ...Bacterial adhesion is a general strategy for host-microbe and microbe-microbe interactions. Adhesive pili are essential for colonization, biofilm formation, virulence and pathogenesis of many environmental and pathogenic bacteria. Members of the class Bacteroidia have unique type V pili, assembled by protease-mediated polymerization. Porphyromonas gingivalis is the main contributor to periodontal disease and its type V pili are a key factor for its virulence. However, the structure of the polymerized pilus and its assembly mechanism are unknown. Here we show structures of polymerized and monomeric states of FimA stalk pilin from P. gingivalis, determined by cryo-electron microscopy and crystallography. The atomic model of assembled FimA shows that the C-terminal strand of a donor subunit is inserted into a groove in the β-sheet of an acceptor subunit after N-terminal cleavage by the protease RgpB. The C terminus of the donor strand is essential for polymerization. We propose that type V pili assemble via a sequential polar assembly mechanism at the cell surface, involving protease-mediated strand exchange, employed by various Gram-negative species belonging to the class Bacteroidia. Our results reveal functional surfaces related to pathogenic properties of polymerized FimA. These insights may facilitate development of antibacterial drugs. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0724.map.gz emd_0724.map.gz | 200.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0724-v30.xml emd-0724-v30.xml emd-0724.xml emd-0724.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0724_fsc.xml emd_0724_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_0724.png emd_0724.png | 43 KB | ||
| Filedesc metadata |  emd-0724.cif.gz emd-0724.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0724 http://ftp.pdbj.org/pub/emdb/structures/EMD-0724 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0724 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0724 | HTTPS FTP |
-Related structure data
| Related structure data |  6kmfMC  6jzjC  6jzkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11015 (Title: Raw data for "Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange" EMPIAR-11015 (Title: Raw data for "Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange"Data size: 72.1 Data #1: FimA motion-corrected dose-weighted frame sums [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0724.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0724.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FimA pilus cryo-EM reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.12 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Type V pilus (FimA)
| Entire | Name: Type V pilus (FimA) |
|---|---|
| Components |
|
-Supramolecule #1: Type V pilus (FimA)
| Supramolecule | Name: Type V pilus (FimA) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Porphyromonas gingivalis ATCC 33277 (bacteria) Porphyromonas gingivalis ATCC 33277 (bacteria) |
| Molecular weight | Theoretical: 6 kDa/nm |
-Macromolecule #1: Major fimbrium subunit FimA type-1
| Macromolecule | Name: Major fimbrium subunit FimA type-1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Porphyromonas gingivalis ATCC 33277 (bacteria) Porphyromonas gingivalis ATCC 33277 (bacteria) |
| Molecular weight | Theoretical: 36.488805 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AFGVGDDESK VAKLTVMVYN GEQQEAIKSA ENATKVEDIK CSAGQRTLVV MANTGAMELV GKTLAEVKAL TTELTAENQE AAGLIMTAE PKTIVLKAGK NYIGYSGTGE GNHIENDPLK IKRVHARMAF TEIKVQMSAA YDNIYTFVPE KIYGLIAKKQ S NLFGATLV ...String: AFGVGDDESK VAKLTVMVYN GEQQEAIKSA ENATKVEDIK CSAGQRTLVV MANTGAMELV GKTLAEVKAL TTELTAENQE AAGLIMTAE PKTIVLKAGK NYIGYSGTGE GNHIENDPLK IKRVHARMAF TEIKVQMSAA YDNIYTFVPE KIYGLIAKKQ S NLFGATLV NADANYLTGS LTTFNGAYTP ANYANVPWLS RNYVAPAADA PQGFYVLEND YSANGGTIHP TILCVYGKLQ KN GADLAGA DLAAAQAANW VDAEGKTYYP VLVNFNSNNY TYDSNYTPKN KIERNHKYDI KLTITGPGTN NPENPITESA HLN VQCTVA EWVLVGQNAT W UniProtKB: Major fimbrium subunit FimA type-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Concentration: 20.0 mM / Component - Formula: C4H11NO3 / Component - Name: Tris |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 100.1 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 1e-06 kPa / Details: Gatan Solarus |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV / Details: 3 second blot, 3.0uL. |
| Details | matured, polymerized state |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 77.0 K / Max: 100.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.1 mrad |
| Details | nanoprobe, parallel beam illumination |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Digitization - Frames/image: 1-75 / Number grids imaged: 1 / Number real images: 1153 / Average exposure time: 60.0 sec. / Average electron dose: 46.0 e/Å2 Details: frame alignment and dose weighting using motioncor2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 30.0 µm / Calibrated magnification: 125000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.5 µm / Nominal defocus min: -1.5 µm / Nominal magnification: 92000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)