+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0706 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of apo Lassa virus polymerase | |||||||||
 Map data Map data | apo lassa virus polymerase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | polymerase / RNA virus / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative stranded viral RNA replication / cap snatching / virion component / host cell / Hydrolases; Acting on ester bonds / host cell cytoplasm / hydrolase activity / RNA-directed RNA polymerase / nucleotide binding / RNA-directed RNA polymerase activity / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Lassa mammarenavirus Lassa mammarenavirus | |||||||||
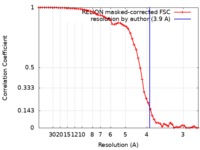
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Peng R / Xu X | |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structural insight into arenavirus replication machinery. Authors: Ruchao Peng / Xin Xu / Jiamei Jing / Min Wang / Qi Peng / Sheng Liu / Ying Wu / Xichen Bao / Peiyi Wang / Jianxun Qi / George F Gao / Yi Shi /  Abstract: Arenaviruses can cause severe haemorrhagic fever and neurological diseases in humans and other animals, exemplified by Lassa mammarenavirus, Machupo mammarenavirus and lymphocytic choriomeningitis ...Arenaviruses can cause severe haemorrhagic fever and neurological diseases in humans and other animals, exemplified by Lassa mammarenavirus, Machupo mammarenavirus and lymphocytic choriomeningitis virus, posing great threats to public health. These viruses encode a large multi-domain RNA-dependent RNA polymerase for transcription and replication of the viral genome. Viral polymerases are one of the leading antiviral therapeutic targets. However, the structure of arenavirus polymerase is not yet known. Here we report the near-atomic resolution structures of Lassa and Machupo virus polymerases in both apo and promoter-bound forms. These structures display a similar overall architecture to influenza virus and bunyavirus polymerases but possess unique local features, including an arenavirus-specific insertion domain that regulates the polymerase activity. Notably, the ordered active site of arenavirus polymerase is inherently switched on, without the requirement for allosteric activation by 5'-viral RNA, which is a necessity for both influenza virus and bunyavirus polymerases. Moreover, dimerization could facilitate the polymerase activity. These findings advance our understanding of the mechanism of arenavirus replication and provide an important basis for developing antiviral therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0706.map.gz emd_0706.map.gz | 1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0706-v30.xml emd-0706-v30.xml emd-0706.xml emd-0706.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0706_fsc.xml emd_0706_fsc.xml | 5.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_0706.png emd_0706.png | 92 KB | ||
| Filedesc metadata |  emd-0706.cif.gz emd-0706.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0706 http://ftp.pdbj.org/pub/emdb/structures/EMD-0706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0706 | HTTPS FTP |
-Related structure data
| Related structure data |  6klcMC  0707C  0708C  0709C  0710C  6kldC  6kleC  6klhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0706.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0706.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | apo lassa virus polymerase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Lassa virus polymerase
| Entire | Name: Lassa virus polymerase |
|---|---|
| Components |
|
-Supramolecule #1: Lassa virus polymerase
| Supramolecule | Name: Lassa virus polymerase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Lassa mammarenavirus Lassa mammarenavirus |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Lassa mammarenavirus Lassa mammarenavirus |
| Molecular weight | Theoretical: 253.505828 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MEEDIACVKD LVSKYLANNE RLSRQKLAFL VQTEPRMLLM EGLKLLSLCI EVDSCNANGC EHNSEDKSVE RILHDHGVLT PSLCFVVPD GYKLTGNVLI LLECFVRSSP ANFEQKYVED FKKLEQLKED LKSVDINLIP LIDGRTSFYN EQIPDWVNDK L RDTLFSLL ...String: MEEDIACVKD LVSKYLANNE RLSRQKLAFL VQTEPRMLLM EGLKLLSLCI EVDSCNANGC EHNSEDKSVE RILHDHGVLT PSLCFVVPD GYKLTGNVLI LLECFVRSSP ANFEQKYVED FKKLEQLKED LKSVDINLIP LIDGRTSFYN EQIPDWVNDK L RDTLFSLL KYAQESNSLF EESEYSRLCE SLSMTSGRLS GVESLNALLD NRSNHYEEVI ASCHQGINNK LTAHEVKLQI EE EYQVFRN RLRKGEIEGQ FLKVEKNQLL NEFNNLYADK VAEKDSVEHL THQFKRASPI LRFLYANISK GDNGEGNLII GEC QMQCWR SFLNKVKSMR ILNTRRKLLL IFDALILLAS KHDQVRKKPL RGWLGTCFVS VNDRLVSLES TKKDLKKWVE RRQQ VERSR TMQSFQCPSK NQILNSIFQK TISKATTALR DVGISVDHYK IDMEVICPDG YDLIMDFDVS GVTPTISYQR SEEEA FPYI MGDVDLLKTT DLERLSSLSL ALVNSMKTSS TVKLRQNEFG PARYQVVKCK EAYCQEFSLG ETEFQLIYQK TGECSK CYA INDNRVGEVC SFYADPKRYF PAIFSAEVLQ TTVSTMISWI EDCNELEEQL DKIRSLTKMI LILILAHPSK RSQKLLQ NL RYFIMAYVSD YYHKDLIDKV REELITDVEF LLYRLLRTLM GLVLSEDVKS MMTNRFKFIL NISYMCHFIT KETPDRLT D QIKCFEKFLE PKVKFGHVSI NPADTATEEE LDDMVYNAKK FLSKGGCTSA KGPSYKKPGV SKKYLSLLTS SFNNGSLFK EREVKKEIKD PLITSGCATA LDLASNKSVV VNKYTDGSRV LNYDFNKLTA LAVSQLTEVF SRKGKHLLNK QDYEYKVQQA MSNLVLGSK QHKGDADEAD LDEILLDGGA STYFNQLKET VEKIVDQYRE PVKMGSGSND GDQPSINDLD EIVSNKFYIR L IKGELSNH MVEDFDHDVL PDKFYEEFCD AVYENSKLKE KYFYCGHMSQ CPIGELTKAV STRTYLDHEY FQCFKSILLI MN ANALMGK YTHYKSRNLN FKFDMGKLSD DARISERESN SEALSKALSL TNCTTAMLKN LCFYSQESPQ SYNSVGPDTG RLK FSLSYK EQVGGNRELY IGDLRTKMFT RLIEDYFEAL SSQLSGSCLN NEKEFENAIL SMKLNVSMAH VSYSMDHSKW GPMM CPFLF LTVLQNLIFL SKDLQADIKG RDYLSTLLMW HMHKMVEIPF NVVSAMMKSF IKAQLGLRKK TKQSITEDFF YSNFQ IGVV PSHISSILDM GQGILHNTSD FYALITERFI NYAISCVCGG TIDAYTSSDD QISLFDQTLT ELLHRDPEEF RALMEF HYY MSDQLNKFVS PKSVIGRFVA EFKSRFFVWG DEVPLLTKFV AAALHNIKCK EPHQLAETID TIVDQSVANG VPVHLCN LI QIRTLSLLQY ARYPIDPFLL NCETDVRDWV DGNRSYRIMR QIEGLIPDAC SKIRSMLRRL YNRLKTGQLH EEFTTNYL S SEHLSSLKNL CELLGVEPPS ESDLEYSWLN LAAHHPLRMV LRQKIIYSGA VNLDDEKIPT IVKTIQNKLS STFTRGAQK LLSEAINKSA FQSSIASGFV GLCRTLGSKC VRGPNKENLY IKSIQSLITG TQGIELLTNS IGVQYWRVPL GLRNKSESVV SYFRPLLWD YMCISLSTAI ELGAWVLGDP KTTKALDFFK HNPCDYFPLK PTASKLLEDR VGLNHIIHSL RRLYPSVFEK H ILPFMSDL ASTKMKWSPR IKFLDLCVAL DVNCEALSLV SHIVKWKREE HYIVLSSELR FSHTRTHEPM VEERVVSTSD AV DNFMRQI YFESYVRPFV ATTRTLGSFT WFPHRTSIPE GEGLHRLGPF SSFVEKVIHK GVERPMFKHD LMMGYAWIDF DIE PARFNQ NQLIASGLVD SKFDSLEDFF DAVASLPTGS AKLSQTVRFR IKSQDASFRE SFAIHLDYIG SMNQQAKYLV HDVT AMYSG AVSPCVLSDC WRLVLSGPTF KGKPVWYVDT EVINEFLVDT NQLGHVTPVE VVVDMEKLQF AEYDFMLVGP CAEPV PLVV RRGGLWECEK KLASFTPVIQ DQDLEMFVRE VGDTSSDLLI RALSDMITDR LGLRMQWSGV DIVSTLRAAA PGNAEV LSA VLEVVDNWVE FKGYALCYSK SRGRVMVQSS SGKLRLKGRT CEELTEGGEH VEDIE UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6klc: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)