+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0632 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rotavirus A-VP3 (RVA-VP3) | ||||||||||||
 Map data Map data | A sub-atomic resolution cryo-EM structure of full-length Rotavirus A-VP3 (RVA-VP3) | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Rotavirus / Capping enzyme / Methyl transferase / RTPase / PDE / STRUCTURAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on ester bonds; Phosphoric-diester hydrolases / viral process / viral nucleocapsid / mRNA guanylyltransferase activity / mRNA guanylyltransferase / mRNA (guanine-N7)-methyltransferase / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / symbiont-mediated suppression of host innate immune response / hydrolase activity / GTP binding / RNA binding Similarity search - Function | ||||||||||||
| Biological species |  Rotavirus A Rotavirus A | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | ||||||||||||
 Authors Authors | Kumar D / Yu X | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: A sub-atomic resolution cryo-EM of full-length Rotavirus A-VP3 (RVA-VP3) Authors: Kumar D / Yu X / Prasad V / Wang Z | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0632.map.gz emd_0632.map.gz | 20.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0632-v30.xml emd-0632-v30.xml emd-0632.xml emd-0632.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0632.png emd_0632.png | 60.8 KB | ||
| Filedesc metadata |  emd-0632.cif.gz emd-0632.cif.gz | 6.2 KB | ||
| Others |  emd_0632_half_map_1.map.gz emd_0632_half_map_1.map.gz emd_0632_half_map_2.map.gz emd_0632_half_map_2.map.gz | 79.4 MB 79.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0632 http://ftp.pdbj.org/pub/emdb/structures/EMD-0632 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0632 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0632 | HTTPS FTP |
-Related structure data
| Related structure data |  6o6bMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0632.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0632.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A sub-atomic resolution cryo-EM structure of full-length Rotavirus A-VP3 (RVA-VP3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
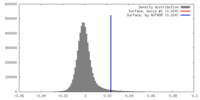
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map of VP3
| File | emd_0632_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of VP3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of VP3
| File | emd_0632_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of VP3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : VP3
| Entire | Name: VP3 |
|---|---|
| Components |
|
-Supramolecule #1: VP3
| Supramolecule | Name: VP3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Rotavirus A Rotavirus A |
-Macromolecule #1: Protein VP3
| Macromolecule | Name: Protein VP3 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO EC number: Hydrolases; Acting on ester bonds; Phosphoric-diester hydrolases |
|---|---|
| Source (natural) | Organism:  Rotavirus A Rotavirus A |
| Molecular weight | Theoretical: 97.70382 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKVLALRHSV AQVYADTQIY THDDTKDSYE NAFLISNLTT HNILYFNYSA RTLEILNKSG IAAIEIQSLE ELFTLIRCNF TYDYENNVV YLHDYSYYTN NEIRTDQHWI TKTNIEEYLL PGWKLTYVGY NGNDTRGHYN FSFTCQNAAT DDDIIIEYIY S EALDFQNF ...String: MKVLALRHSV AQVYADTQIY THDDTKDSYE NAFLISNLTT HNILYFNYSA RTLEILNKSG IAAIEIQSLE ELFTLIRCNF TYDYENNVV YLHDYSYYTN NEIRTDQHWI TKTNIEEYLL PGWKLTYVGY NGNDTRGHYN FSFTCQNAAT DDDIIIEYIY S EALDFQNF MLRKIKERMT TSLPIARLSN RVFRDKLFPL LVKKHKRVVN VGPRNESMFT FLNFPSIRQF SNGPYLVKNT IK LKQERWL GKRVSQFDIG QYKNMLNVIT TIYHYYNLYQ EKPIIYMVGS APSYWIYDVR QYSDFLFETW DPLDTPYSSI HHK ELFFAK DIGKLKDNSI LYIDIRTDRG NADWKEWRKV VELQTISNLN LAYQYLATGK SKVCCVKLTA MDLELPVSAK LLHH PTTEI RSEFYLLLDI WDVNNIKRFI PKGALYSFIN NVITDNVFIQ SPFKIRTSVS DYIVALYALS NDFNNREDII NLINN QKQS LITVRINNTF KDEPKVGFKS IYDWTFLPTD FETTNAIVTS YDGCLGIFGL SISLASKPTG NNHLFILNGT DKYYKL DQF ANHTGISRRS HQVRFSESAT SYSGYIFRDL SNSNFNLIGT NVENSVSGHV YNALIYYRYN YSFDLKRWIY LHSVEKA NI EGGKYYEHAP IELIYACKSA KEFASLQDDL TVLRYANEIE NYINKVYSIT YADDPNYFIG IKFNNIPYIY DVKVPHLT F GVLYISDNMI PDVVKIMKSM KQELFGMDVT TSYTYMLSDG VYVANVSGVL ATYFKMYNLF YKNQITFGQS RMFIPHITL SFSNNKTVRI ETTKLRIKSI YLRKIRGDTV FDMPE UniProtKB: Protein VP3 |
-Macromolecule #2: GUANOSINE-5'-MONOPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-MONOPHOSPHATE / type: ligand / ID: 2 / Number of copies: 4 / Formula: 5GP |
|---|---|
| Molecular weight | Theoretical: 363.221 Da |
| Chemical component information |  ChemComp-5GP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7420 pixel / Digitization - Dimensions - Height: 7676 pixel / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 40000 |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)