+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0272 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | F97L Hepatitis B core protein capsid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Hapatitis B core protein Hbc Hbcag / VIRUS LIKE PARTICLE premature envelopment mutant F97L / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule-dependent intracellular transport of viral material towards nucleus / T=4 icosahedral viral capsid / viral penetration into host nucleus / host cell / host cell cytoplasm / symbiont entry into host cell / structural molecule activity / DNA binding / RNA binding Similarity search - Function | |||||||||
| Biological species |   Hepatitis B virus Hepatitis B virus | |||||||||
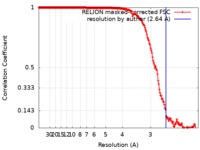
| Method | single particle reconstruction / cryo EM / Resolution: 2.64 Å | |||||||||
 Authors Authors | Bottcher B / Nassal M | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2018 Journal: J Mol Biol / Year: 2018Title: Structure of Mutant Hepatitis B Core Protein Capsids with Premature Secretion Phenotype. Authors: Bettina Böttcher / Michael Nassal /  Abstract: Hepatitis B virus is a major human pathogen that consists of a viral genome surrounded by an icosahedrally ordered core protein and a polymorphic, lipidic envelope that is densely packed with surface ...Hepatitis B virus is a major human pathogen that consists of a viral genome surrounded by an icosahedrally ordered core protein and a polymorphic, lipidic envelope that is densely packed with surface proteins. A point mutation in the core protein in which a phenylalanine at position 97 is exchanged for a smaller leucine leads to premature envelopment of the capsid before the genome maturation is fully completed. We have used electron cryo-microscopy and image processing to investigate how the point mutation affects the structure of the capsid at 2.6- to 2.8 Å-resolution. We found that in the mutant the smaller side chain at position 97 is displaced, increasing the size of an adjacent pocket in the center of the spikes of the capsid. In the mutant, this pocket is filled with an unknown density. Phosphorylation of serine residues in the unresolved C-terminal domain of the mutant leaves the structure of the ordered capsid largely unchanged. However, we were able to resolve several previously unresolved residues downstream of proline 144 that precede the phosphorylation-sites. These residues pack against the neighboring subunits and increase the inter-dimer contact suggesting that the C-termini play an important role in capsid stabilization and provide a much larger interaction interface than previously observed. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0272.map.gz emd_0272.map.gz | 480.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0272-v30.xml emd-0272-v30.xml emd-0272.xml emd-0272.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0272_fsc.xml emd_0272_fsc.xml | 18 KB | Display |  FSC data file FSC data file |
| Images |  emd_0272.png emd_0272.png | 428.5 KB | ||
| Masks |  emd_0272_msk_1.map emd_0272_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0272.cif.gz emd-0272.cif.gz | 6.4 KB | ||
| Others |  emd_0272_half_map_1.map.gz emd_0272_half_map_1.map.gz emd_0272_half_map_2.map.gz emd_0272_half_map_2.map.gz | 409.1 MB 409.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0272 http://ftp.pdbj.org/pub/emdb/structures/EMD-0272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0272 | HTTPS FTP |
-Related structure data
| Related structure data |  6hu4MC  0271C  0273C  6htxC  6hu7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0272.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0272.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.053 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
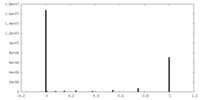
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0272_msk_1.map emd_0272_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
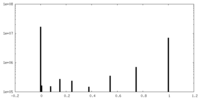
| Density Histograms |
-Half map: #1
| File | emd_0272_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
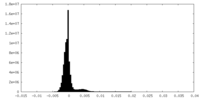
| Density Histograms |
-Half map: #2
| File | emd_0272_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
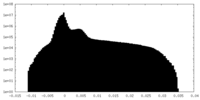
| Density Histograms |
- Sample components
Sample components
-Entire : Hepatitis B virus
| Entire | Name:   Hepatitis B virus Hepatitis B virus |
|---|---|
| Components |
|
-Supramolecule #1: Hepatitis B virus
| Supramolecule | Name: Hepatitis B virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10407 / Sci species name: Hepatitis B virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:   Hepatitis B virus / Strain: ayw Hepatitis B virus / Strain: ayw |
| Molecular weight | Theoretical: 4.8 MDa |
| Virus shell | Shell ID: 1 / Name: WT Hbc / Diameter: 340.0 Å / T number (triangulation number): 4 |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 Details: Hepatitis B core protein premature envelopment mutant F97L Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Hepatitis B virus Hepatitis B virus |
| Molecular weight | Theoretical: 21.112199 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDIDPYKEFG ATVELLSFLP SDFFPSVRDL LDTASALYRE ALESPEHCSP HHTALRQAIL CWGELMTLAT WVGVNLEDPA SRDLVVSYV NTNMGLKLRQ LLWFHISCLT FGRETVIEYL VSFGVWIRTP PAYRPPNAPI LSTLPETTVV RRRGRSPRRR T PSPRRRRS QSPRRRRSQS RESQC UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.7 Component:
Details: 25 mM Tris pH 7.7, 150 mM NaCl | |||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV Details: Samples were vitrified with an FEI Vitrobot IV, which was operated at 100% humidity at 4C. 2.5-3.5 ul of sample were applied to grids (Quantifoil UltrAuFoil R1.3/1.2 300 mesh gold grids ) ...Details: Samples were vitrified with an FEI Vitrobot IV, which was operated at 100% humidity at 4C. 2.5-3.5 ul of sample were applied to grids (Quantifoil UltrAuFoil R1.3/1.2 300 mesh gold grids ) that were glow discharged in air for 1-2 min. The sample was incubated for 30 s before blotting for 3 s with a blot force between 0 and 5.. | |||||||||
| Details | sample is purified from e.coli via sucrose density gradient. sucrose is removed via buffer exchange on a concentrator |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-28 / Number grids imaged: 1 / Number real images: 1948 / Average electron dose: 28.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.56 µm / Calibrated defocus min: 0.37 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)